 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI06G00310.4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 348aa MW: 38340.5 Da PI: 6.1271 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 26.3 | 1.8e-08 | 115 | 153 | 1 | 41 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqck 41
r +WT E++++++a +++ + Wk+I +++g +t q++
OMERI06G00310.4 115 RESWTDPEHDKFLEALQLFDRD-WKKIEAYVG-SKTVIQLV 153
789*****************77.*********.99998875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.26E-10 | 103 | 155 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.4E-9 | 113 | 151 | IPR006447 | Myb domain, plants |
| PROSITE profile | PS51293 | 10.009 | 113 | 164 | IPR017884 | SANT domain |
| SMART | SM00717 | 9.8E-7 | 114 | 162 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.5E-4 | 114 | 151 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 5.0E-6 | 115 | 151 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.97E-5 | 117 | 160 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0032922 | Biological Process | circadian regulation of gene expression | ||||
| GO:0043966 | Biological Process | histone H3 acetylation | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MEVRRKGVWG PSVSVSGGTR GQEKNPRCDV FLLTPRSAPF SSSSSSSPPA NHLTKQAVEL 60 STPTQPNPRL RLCEIAGMSS PPQQLLDSSS PGPGPGVEDD GGRRVRKPYT ITKSRESWTD 120 PEHDKFLEAL QLFDRDWKKI EAYVGSKTVI QLVAFVTEYV TLLQFLNLAG IDLIINLGVL 180 AAPQVVLPQQ ASHLMEQGCL IPMDISPVAR NFNANDVFSS WDSAIAQSFS PRHTHESQSG 240 TCPTSEAIEQ EIMLPTLRAM PDFAQVYNFL GSIFDPETSG HLQRLREMDP IDVETVLLLM 300 KNLSINLTNP NFEAHRKVFA SHGSGMDQVK HESLGDLGST HTLHLPFM |
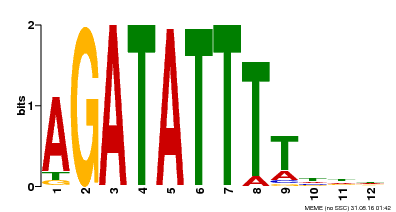
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00338 | DAP | Transfer from AT3G09600 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK111672 | 1e-175 | AK111672.1 Oryza sativa Japonica Group cDNA clone:J013156D15, full insert sequence. | |||
| GenBank | AK119640 | 1e-175 | AK119640.1 Oryza sativa Japonica Group cDNA clone:002-130-F07, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015642931.1 | 1e-158 | protein REVEILLE 6 | ||||
| TrEMBL | A0A0E0DVJ5 | 0.0 | A0A0E0DVJ5_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI06G00320.1 | 0.0 | (Oryza meridionalis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09600.1 | 2e-58 | MYB_related family protein | ||||



