 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI06G09270.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 385aa MW: 40556.4 Da PI: 6.5612 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 55.3 | 9.2e-18 | 262 | 296 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C +C t kTp+WR gp g+ktLCnaCG++y++ +l
OMERI06G09270.1 262 CLHCETDKTPQWRTGPMGPKTLCNACGVRYKSGRL 296
99*****************************9885 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF016992 | 1.0E-71 | 15 | 345 | IPR016679 | Transcription factor, GATA, plant |
| Gene3D | G3DSA:3.30.50.10 | 4.4E-15 | 256 | 293 | IPR013088 | Zinc finger, NHR/GATA-type |
| SMART | SM00401 | 1.3E-18 | 256 | 306 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 12.453 | 256 | 292 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 1.24E-15 | 258 | 319 | No hit | No description |
| CDD | cd00202 | 1.56E-14 | 261 | 312 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 262 | 287 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 1.4E-15 | 262 | 296 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 385 aa Download sequence Send to blast |
MEVTAEFGGA YYGGAAGREK KALQQGCGDH FAVDDLLVLP YDEEDETTRE GEATGGKEEA 60 AGFGNASADS STITALDSCS NSFGLADGDF PGELCEPYDQ LAELEWLSNY MNDGDDAFAT 120 EDLQKLHLIS GIPFGGFSTS SVPSAQAASA AASMAVQPGG FLPEAPVPAK ARSKRSRAAP 180 GNWSSRLLVL PPPPASPPSP ASMAISPAES GVSAHAFPIK KPSKPAKKKD APAPPAQAQL 240 SSVPVHSGGS APAAAAGEGR RCLHCETDKT PQWRTGPMGP KTLCNACGVR YKSGRLVPEY 300 RPAASPTFMV SKHSNSHRKV LELRRQKEMH QQTPHHHQPQ VAAAGGVGSL MHMQSSMLFD 360 GVSPVVSGDD FLIHHHLRTD FRPLI |
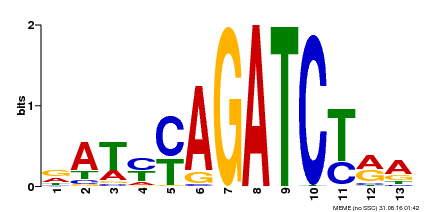
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00530 | DAP | Transfer from AT5G25830 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK121896 | 0.0 | AK121896.1 Oryza sativa Japonica Group cDNA clone:J033105E11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631175.1 | 0.0 | GATA transcription factor 12 | ||||
| TrEMBL | A0A0E0DZ74 | 0.0 | A0A0E0DZ74_9ORYZ; GATA transcription factor | ||||
| STRING | OMERI06G08780.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3025 | 31 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G25830.1 | 7e-50 | GATA transcription factor 12 | ||||



