 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI07G16820.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 614aa MW: 65773.1 Da PI: 6.3317 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101.2 | 6.2e-32 | 233 | 290 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C vkk +ers d++++e++Y+g+Hnh+k
OMERI07G16820.1 233 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCDVKKLLERSL-DGQITEVVYKGRHNHPK 290
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.4 | 1.5e-33 | 405 | 463 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct +gCpv+k+ver+++dpk v++tYeg+Hnhe
OMERI07G16820.1 405 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNTGCPVRKHVERASHDPKSVITTYEGKHNHE 463
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.2E-27 | 226 | 291 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.789 | 227 | 291 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.89E-25 | 231 | 291 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.3E-35 | 232 | 290 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.5E-24 | 233 | 289 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-37 | 390 | 465 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-29 | 397 | 465 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.526 | 400 | 465 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.7E-39 | 405 | 464 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.4E-26 | 406 | 463 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 614 aa Download sequence Send to blast |
MADSPNPSSG DHPAGVGGSP EKQPPVDRRV AALAAGAGAR YKAMSPARLP ISREPCLTIP 60 AGFSPSALLE SPVLLTNFKV EPSPTTGTLS MAAIMNKSAN PDILPSPRDK SSGSTHEDGG 120 SRDFEFKPHL NSSSQSTAPA INDPKKHETS MKNESLNTAP SSDDMMIDNI PLCSRESTLA 180 VNVSSAPSQP VGMVGLTDSA PAEVGASELH QMNSSGNAMQ ESQPESVAEK SAEDGYNWRK 240 YGQKHVKGSE NPRSYYKCTH PNCDVKKLLE RSLDGQITEV VYKGRHNHPK PQPNRRLSAG 300 AVPPSQGEER YDVATTDDKS SNVLSNLGNA VHTAGMIEPV PGTASDDDND AGGGRPYPGD 360 DAVEDDDLES KRRKMESAAI DAALMGKPNR EPRVVVQTVS EVDILDDGYR WRKYGQKVVK 420 GNPNPRSYYK CTNTGCPVRK HVERASHDPK SVITTYEGKH NHEVPASRNA SHEMSTPPMK 480 PVVHPINSNM QGLGGMMRAC EPRTFPNQYS QAAESDTISL DLGVGISPNH SDATNQLQSS 540 VSDQMQYQMQ PMGSVYSNMG LPAMAMPTMA GNAASSIYGS REEKPSEGFT FKATPMDHSA 600 NLCYSTAGNL VMGP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 8e-37 | 233 | 466 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 8e-37 | 233 | 466 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
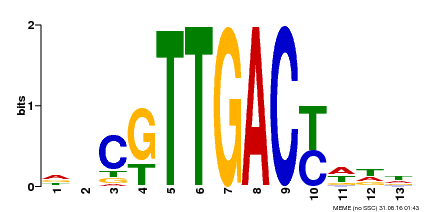
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK070537 | 0.0 | AK070537.1 Oryza sativa Japonica Group cDNA clone:J023062H03, full insert sequence. | |||
| GenBank | AY341850 | 0.0 | AY341850.1 Oryza sativa (japonica cultivar-group) WRKY9 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646742.1 | 0.0 | WRKY transcription factor SUSIBA2 isoform X1 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | A0A0E0EDN9 | 0.0 | A0A0E0EDN9_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI07G17160.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6740 | 38 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-131 | WRKY family protein | ||||



