 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI08G18590.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 378aa MW: 40384.5 Da PI: 8.038 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 22.4 | 3.2e-07 | 201 | 223 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cgk F+r nL+ H+r H
OMERI08G18590.1 201 HYCQVCGKGFKRDANLRMHMRAH 223
68*******************99 PP
| |||||||
| 2 | zf-C2H2 | 11.9 | 0.00069 | 289 | 312 | 1 | 23 |
EEETTTT.EEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCg.ksFsrksnLkrHirtH 23
y+C+ Cg k+Fs s+L++H + +
OMERI08G18590.1 289 YVCNRCGrKHFSVLSDLRTHEKHC 312
9*******************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.99E-6 | 199 | 226 | No hit | No description |
| PROSITE profile | PS50157 | 12.653 | 201 | 228 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0034 | 201 | 223 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.7E-7 | 201 | 230 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 203 | 223 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 120 | 251 | 284 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 35 | 289 | 315 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MIPGGGGGGG GGGGVHGGGG ISPYLVQSQH GRGGGVDGME MEEGGGYMGE QAQCHPLLYN 60 LSVLKDRVQQ LHPLVGLVVA HNAHAHGPLD VSAADAIIQE IVAAASSMMY AFQLLCDLGT 120 APTTAPPQET AVVVKNNDDH AGQMEDDHLM QQQWQQNGSR QHDYSSHAPP VFHGETAAGA 180 TSATDTIIEL DAAELLAKYT HYCQVCGKGF KRDANLRMHM RAHGDEYKSK AALSNPSKGG 240 DEMMAAAARK YSCPQEGCRW NRGHAKFQPL KSVICAKNHY KRSHCPKMYV CNRCGRKHFS 300 VLSDLRTHEK HCGNHRWLCS CGTSFSRKDK LIGHVSLFAG HQPVMPLDAP RAGKRQRSSS 360 ASVAGNIDDT TGIGMGAA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Together with STOP1, plays a critical role in tolerance to major stress factors in acid soils such as proton H(+) and aluminum ion Al(3+). Required for the expression of genes in response to acidic stress (e.g. ALMT1 and MATE), and Al-activated citrate exudation. {ECO:0000269|PubMed:23935008}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00522 | DAP | Transfer from AT5G22890 | Download |
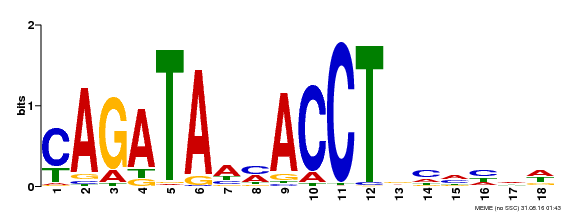
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK063290 | 0.0 | AK063290.1 Oryza sativa Japonica Group cDNA clone:001-113-E02, full insert sequence. | |||
| GenBank | AP004587 | 0.0 | AP004587.3 Oryza sativa Japonica Group genomic DNA, chromosome 8, PAC clone:P0543D10. | |||
| GenBank | AP005544 | 0.0 | AP005544.3 Oryza sativa Japonica Group genomic DNA, chromosome 8, PAC clone:P0604E01. | |||
| GenBank | AP014964 | 0.0 | AP014964.1 Oryza sativa Japonica Group DNA, chromosome 8, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015649514.1 | 0.0 | protein SENSITIVE TO PROTON RHIZOTOXICITY 2 | ||||
| Swissprot | Q0WT24 | 4e-94 | STOP2_ARATH; Protein SENSITIVE TO PROTON RHIZOTOXICITY 2 | ||||
| TrEMBL | A0A0E0EP39 | 0.0 | A0A0E0EP39_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI08G18460.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7441 | 32 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22890.1 | 1e-94 | C2H2 family protein | ||||



