 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI09G14420.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 535aa MW: 55113.4 Da PI: 9.5117 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.4 | 1.3e-05 | 79 | 101 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ C+k F r nL+ H r H
OMERI09G14420.1 79 FVCEVCNKGFQREQNLQLHRRGH 101
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 12.5 | 0.00043 | 155 | 177 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C +C+k++ +s+ k H +t+
OMERI09G14420.1 155 WRCDKCSKRYAVQSDWKAHSKTC 177
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 7.9E-7 | 78 | 101 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 7.3E-6 | 78 | 101 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.094 | 79 | 101 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF12171 | 3.4E-5 | 79 | 101 | IPR022755 | Zinc finger, double-stranded RNA binding |
| SMART | SM00355 | 0.0041 | 79 | 101 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 81 | 101 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.0E-4 | 143 | 176 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 7.9E-7 | 150 | 175 | No hit | No description |
| SMART | SM00355 | 150 | 155 | 175 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 535 aa Download sequence Send to blast |
MAAASSAPLF GLGDTQMPPP QNPTNPALHH HPNPSPAPVV AAAAPAPKKK RNQPGNPSKY 60 PDAEVIALSP RTLMATNRFV CEVCNKGFQR EQNLQLHRRG HNLPWKLKQK NPKEARRRVY 120 LCPEPSCVHH DPSRALGDLT GIKKHYCRKH GEKKWRCDKC SKRYAVQSDW KAHSKTCGTR 180 EYRCDCGTLF SRRDSFITHR AFCDALAQEN ARMPPIGAGV YGGAGNMTLG LTGMAAPQLP 240 AGFTDQAGQA PSASAGDVLR LGGGSNGASQ FDHLMASSSG SSMFRSQGSS SSSFYLANAA 300 SHHAPAQDFG PEDGQSQAGQ GSLLHGKPAA FHDLMQLPVQ HQQSSNGNLL NLGFFSGSNG 360 GVDQFNGGAG NGGQGSIVTS SGLAGNHGGG GGGFPSLYNS SEPAGTLPQM SATALLQKAA 420 QMGATTSSYN AGGSSLLRGA SSHGITAGEG PANERSSYQN LIMGSMASGG GGAGFAGSFS 480 GASGFGGAVD DGKLSTRDFL GVGVVQGISG SPAMGPPRHG AAGLHVGSLD PANMN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 2e-35 | 151 | 213 | 3 | 65 | Zinc finger protein JACKDAW |
| 5b3h_F | 2e-35 | 151 | 213 | 3 | 65 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a positive regulator of the starch synthase SS4. Controls chloroplast development and starch granule formation (PubMed:22898356). Binds DNA via its zinc fingers (PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). {ECO:0000269|PubMed:22898356, ECO:0000269|PubMed:24821766}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
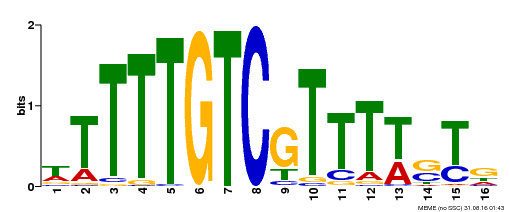
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK101181 | 0.0 | AK101181.1 Oryza sativa Japonica Group cDNA clone:J033029F02, full insert sequence. | |||
| GenBank | AK101950 | 0.0 | AK101950.1 Oryza sativa Japonica Group cDNA clone:J033075E02, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015612461.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_015612462.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_015612463.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_015612465.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Refseq | XP_015612466.1 | 0.0 | protein indeterminate-domain 5, chloroplastic | ||||
| Swissprot | Q9ZUL3 | 1e-120 | IDD5_ARATH; Protein indeterminate-domain 5, chloroplastic | ||||
| TrEMBL | A0A0E0EUR8 | 0.0 | A0A0E0EUR8_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI09G14530.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1733 | 37 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02080.1 | 1e-112 | indeterminate(ID)-domain 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



