 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI12G12160.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 1644aa MW: 187191 Da PI: 9.1846 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 45.2 | 1.7e-14 | 59 | 133 | 18 | 98 |
--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 18 lpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
+p kfa+ + g+ s ++le ++g++++v++ k +++ vl++GW F+++ +Lk+ D++vF+++g+s+f +v++f+
OMERI12G12160.2 59 VPTKFANIIRGQ--ISEVVKLEVPNGKTYDVQV--AKEQNELVLRSGWGAFARDYELKQCDILVFTYSGSSRF--KVRIFN 133
799999666766..6779***************..*********************************99999..999987 PP
| |||||||
| 2 | B3 | 49.6 | 7.3e-16 | 183 | 263 | 12 | 98 |
HTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 12 ksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
+ +++pk fa++ +g+ s +le ++gr+++v++ k ++ v+++GW+ F++a +L++gD++vF ++g+s f +v++f+
OMERI12G12160.2 183 LDKPVTIPKMFAKNVQGQ--ISGVAKLEVPDGRTYDVEI--AKEHNELVFRSGWEVFASAYELEQGDILVFAYSGNSHF--KVQIFN 263
555689******888888..6669***************..**********************************9999..999987 PP
| |||||||
| 3 | B3 | 54.9 | 1.6e-17 | 370 | 461 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvv 94
ffkv+++ + k l +pkkf+++ gk + +++le ++g+ ++v++ + +++ vl++GW +F++ +Lk gD +vF ++g+s f +v
OMERI12G12160.2 370 FFKVMMSVSDIKDE-LAIPKKFTANVRGK--IPEQVRLEVSDGKMYNVQV--TEEQDELVLRSGWANFASTYQLKHGDLLVFIHSGHSHF--KV 456
99***999999987.********888666..3446***************..********************************999999..99 PP
EEE-S CS
B3 95 kvfrk 99
+f+
OMERI12G12160.2 457 LIFDP 461
99885 PP
| |||||||
| 4 | B3 | 35.7 | 1.6e-11 | 587 | 666 | 12 | 92 |
HTT-EE--HHH.HTT---..--SEEEEEETTS.-EEEEEE..EEETTE.EEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE.. CS
B3 12 ksgrlvlpkkfaeehggkkeesktltledesg.rsWevkliyrkksgr.yvltkGWkeFvkangLkegDfvvFkldgr.sefel 92
+sg lv++k++a ++ ++ +++t++l ++ g ++W++ l + + y l++GW +F + n L+egD++vF+ +++ ++ l
OMERI12G12160.2 587 ASGSLVFSKDYAVRYLLD--QNRTIKLCQSGGsKTWDISL--DMDTDDlYALSTGWLDFFRGNLLQEGDICVFEASKSkRGVAL 666
45679**********888..4558888887777*******..8888888*************************8876444444 PP
| |||||||
| 5 | B3 | 38.1 | 2.7e-12 | 710 | 796 | 13 | 99 |
TT-EE--HHH.HTT---..--SEEEEEE.TTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE..EEEEE-S CS
B3 13 sgrlvlpkkfaeehggkkeesktltled.esgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr.sefelvvkvfrk 99
+ r +++ k+a+++ e++ ++l+ e+ + + l+++ + + + l kGW +Fv++n+L++ D+++F+l+++ ++ +++v+++rk
OMERI12G12160.2 710 QERKCFSVKYASKYLPH--EDQNMRLRLpETKYKCKAALRVDTSTNLHKLLKGWGKFVNDNKLEVHDICLFQLMKNkKKLTMTVHIIRK 796
45677888999999433..3445777763455778888877999999***************************999*********997 PP
| |||||||
| 6 | B3 | 57.3 | 2.9e-18 | 836 | 923 | 1 | 97 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvv 94
f+kv+ +k+g +++p kfa+++gg+ s t++le+++g+++ev++ k + vl++GW++F++a +L+ gD++vF +g+s f +v
OMERI12G12160.2 836 FIKVM--ITDFKNG-VTIPAKFARNFGGQM--SGTVKLETRNGKTYEVQV--AKELNNLVLRSGWERFASAYELEKGDILVFIQSGNSHF--KV 920
67777..6678887.********9999994..559***************..*******************************8888888..66 PP
EEE CS
B3 95 kvf 97
++
OMERI12G12160.2 921 WIY 923
665 PP
| |||||||
| 7 | B3 | 40.3 | 5.8e-13 | 1052 | 1133 | 8 | 93 |
-HHHHTT-EE--HHH.HTT---..--SEEEEEE.TTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..E CS
B3 8 sdvlksgrlvlpkkfaeehggkkeesktltled.esgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelv 93
+ + + g+l ++k++a ++ ++k+e t++l + ++ +W++ l ++ +++y l++GW +F + n+L+egD++vF+ ++++++ v
OMERI12G12160.2 1052 KASATDGLLAISKDYAVSYLLDKNE--TIKLCHsGRSLTWDIGLDIDT-DDQYALSTGWLDFIRNNHLQEGDICVFEASKNKRG--V 1133
55677899***********888767..56665537889******4333.346*************************8887666..3 PP
| |||||||
| 8 | B3 | 37.6 | 4e-12 | 1210 | 1290 | 13 | 94 |
TT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE..EE CS
B3 13 sgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr.sefelvv 94
s+ ++++ k+a+++ + e++ +++ + +s++ k ++ + + + l++GW +Fv++n+Lk gD+++F+l+++ ++ +++v
OMERI12G12160.2 1210 SRYMCFSAKYAAKYL-PRENN-KIMRLRLPKKSYKYKAVFKINNKVHKLGGGWGKFVDDNKLKLGDICLFQLMKDkKKLTMMV 1290
556788889999993.33334.455555566777777777999999***************************9867776666 PP
| |||||||
| 9 | B3 | 42.8 | 9.3e-14 | 1328 | 1404 | 14 | 94 |
T-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EE CS
B3 14 grlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvv 94
+ +++p k ae ++g+ ++ l+++sg++W+v + +k ++ +l +GW++F+ka++L+e+D + F+ +g+ ++++ +
OMERI12G12160.2 1328 HGISIPEKVAEIFSGQITK--GFNLKSPSGETWRVGI--EKVADELILMSGWEDFAKAHELQENDLLFFTCNGHGNGSCSF 1404
459*******777777555..59999***********..*******************************77665554443 PP
| |||||||
| 10 | B3 | 44.4 | 2.9e-14 | 1537 | 1633 | 1 | 97 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEE.TTS-EEEEEE...EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltled.esgrsWevkliy.rkksgryvltkGWkeFvkangLkegDfvvFkldgr.se 89
f+ vl +++v ++++l++p +fa++h + +s ++ l + ++W+vk+ y +++ +++ W +F ++n+L+eg +++F+l++ ++
OMERI12G12160.2 1537 FVAVLQTAHVRRRNILIVPTRFAADHLER--KSHDILLIRpNRKQKWSVKYYYlSNTTRGFNCH-RWIKFIRENRLREGNVCIFELMKGaRR 1625
7889999*******************544..45567776636779******8865555555544.8*******************9976688 PP
E..EEEEE CS
B3 90 felvvkvf 97
+++v+v+
OMERI12G12160.2 1626 PTMTVHVI 1633
88888775 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 2.3E-18 | 23 | 137 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 2.8E-6 | 55 | 135 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 2.55E-18 | 55 | 137 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 11.293 | 59 | 135 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 7.45E-17 | 59 | 133 | No hit | No description |
| Pfam | PF02362 | 9.1E-12 | 59 | 133 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.224 | 172 | 265 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 8.3E-9 | 173 | 265 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 8.9E-19 | 175 | 267 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.35E-18 | 176 | 267 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 1.0E-13 | 182 | 263 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 8.59E-17 | 188 | 263 | No hit | No description |
| SuperFamily | SSF101936 | 1.53E-20 | 363 | 462 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 7.2E-21 | 365 | 462 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 4.76E-17 | 368 | 460 | No hit | No description |
| PROSITE profile | PS50863 | 12.76 | 369 | 462 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.8E-15 | 370 | 460 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.2E-10 | 370 | 462 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.7E-13 | 570 | 673 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.29E-14 | 571 | 674 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 11.124 | 576 | 674 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 3.7E-4 | 578 | 674 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.9E-9 | 587 | 666 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.02E-9 | 588 | 672 | No hit | No description |
| Gene3D | G3DSA:2.40.330.10 | 2.8E-13 | 695 | 797 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.31E-15 | 697 | 799 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 0.0015 | 699 | 797 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.5E-10 | 709 | 796 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.58E-10 | 710 | 795 | No hit | No description |
| PROSITE profile | PS50863 | 8.67 | 757 | 797 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 1.63E-21 | 830 | 928 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.5E-21 | 832 | 928 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 13.084 | 833 | 926 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.81E-19 | 834 | 924 | No hit | No description |
| Pfam | PF02362 | 3.4E-16 | 836 | 922 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.4E-15 | 836 | 926 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 2.2E-15 | 1039 | 1137 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.47E-16 | 1039 | 1138 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 12.111 | 1045 | 1140 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.4E-5 | 1045 | 1143 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.0E-9 | 1053 | 1132 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 4.09E-10 | 1060 | 1125 | No hit | No description |
| Gene3D | G3DSA:2.40.330.10 | 9.5E-16 | 1189 | 1295 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 9.81E-18 | 1190 | 1296 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 3.95E-15 | 1195 | 1290 | No hit | No description |
| SMART | SM01019 | 0.0015 | 1197 | 1294 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 10.165 | 1198 | 1294 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 3.0E-10 | 1209 | 1290 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 5.3E-17 | 1311 | 1414 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 12.167 | 1315 | 1412 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 2.81E-13 | 1317 | 1410 | No hit | No description |
| SMART | SM01019 | 3.3E-7 | 1318 | 1412 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 3.3E-16 | 1318 | 1414 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 5.2E-11 | 1327 | 1402 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.9E-17 | 1530 | 1635 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 9.02E-20 | 1531 | 1631 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.29E-17 | 1535 | 1631 | No hit | No description |
| PROSITE profile | PS50863 | 13.155 | 1537 | 1636 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 3.9E-12 | 1537 | 1633 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 6.0E-10 | 1537 | 1637 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005773 | Cellular Component | vacuole | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1644 aa Download sequence Send to blast |
MRKSFTTCKE CIFYHYWNHM GDQKRSFINV MIGDFVPNAL HEVHSKLDVG LIWLFLQAVP 60 TKFANIIRGQ ISEVVKLEVP NGKTYDVQVA KEQNELVLRS GWGAFARDYE LKQCDILVFT 120 YSGSSRFKVR IFNPSGCEKE LSCVMMNSTP CGHEGSMPYH DNHVQSPSER SSQHATVLPP 180 VLLDKPVTIP KMFAKNVQGQ ISGVAKLEVP DGRTYDVEIA KEHNELVFRS GWEVFASAYE 240 LEQGDILVFA YSGNSHFKVQ IFNPSNCEKE LSCVMMNRSI SDDNHRQSPS WESLVPASRR 300 RRSPPACRLT APLPASQHAA YLQAAPPPAT AVRINVALKF IIVMNKSGTT CMDCITNHYW 360 LHMDDRERYF FKVMMSVSDI KDELAIPKKF TANVRGKIPE QVRLEVSDGK MYNVQVTEEQ 420 DELVLRSGWA NFASTYQLKH GDLLVFIHSG HSHFKVLIFD PSYTEKEFSC VVTDSTSHVH 480 ERSISHDNHL QSPRSVILGK NYSLCSSRKR SRMNPADYPS QRPDVPSSED IKDPMSSGGL 540 QKSKKSCYVL PMLYSMTSAQ EAEVLALEKK IQPQIPLYIT AMDISVASGS LVFSKDYAVR 600 YLLDQNRTIK LCQSGGSKTW DISLDMDTDD LYALSTGWLD FFRGNLLQEG DICVFEASKS 660 KRGVALTFHP FKESHCPKSS EYTLSTKSPT RRVPKRDYFA TNLTNLTDQQ ERKCFSVKYA 720 SKYLPHEDQN MRLRLPETKY KCKAALRVDT STNLHKLLKG WGKFVNDNKL EVHDICLFQL 780 MKNKKKLTMT VHIIRKGECS GSLCCRCCGM EKSHRVCKNC VANHYWLHMD NHGKSFIKVM 840 ITDFKNGVTI PAKFARNFGG QMSGTVKLET RNGKTYEVQV AKELNNLVLR SGWERFASAY 900 ELEKGDILVF IQSGNSHFKV WIYDPSACEK ELPCIITEQL PRVQQRSISH DNHTRLKRNA 960 KSAKLYVDSS GHTKETSEIN PASSPSWKPT ERVPFSEELD EPVDLANVQK ATKSFYSLPR 1020 MCNLTSAQKA EVDALEKRIK PQIPFYITVI DKASATDGLL AISKDYAVSY LLDKNETIKL 1080 CHSGRSLTWD IGLDIDTDDQ YALSTGWLDF IRNNHLQEGD ICVFEASKNK RGVALIFHPL 1140 KQSHHPKPPG CVPSTKFPRH GVSKPNYIVS RFTTLSGQLK IKVEAKVQAI QSEVPIFVAV 1200 MRESFIRGRS RYMCFSAKYA AKYLPRENNK IMRLRLPKKS YKYKAVFKIN NKVHKLGGGW 1260 GKFVDDNKLK LGDICLFQLM KDKKKLTMMV MGEKGCESCR KWQEHYYKEH MDVSRIRFFR 1320 LMTGDFAHGI SIPEKVAEIF SGQITKGFNL KSPSGETWRV GIEKVADELI LMSGWEDFAK 1380 AHELQENDLL FFTCNGHGNG SCSFDVLIFD ASGCEKVSCF FTGKKNSYMC KNFNSIGGQI 1440 AGKYLSSDSE DTSTPSVLIG SPHKASTSKK LSGKTKTNPR KEPEDPNCSH WHVIEEKNTD 1500 DDEHADYHYT RFANYLTGEE RDEIFSLVSL QPGNPVFVAV LQTAHVRRRN ILIVPTRFAA 1560 DHLERKSHDI LLIRPNRKQK WSVKYYYLSN TTRGFNCHRW IKFIRENRLR EGNVCIFELM 1620 KGARRPTMTV HVICKADNRF VLLS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00468 | DAP | Transfer from AT4G33280 | Download |
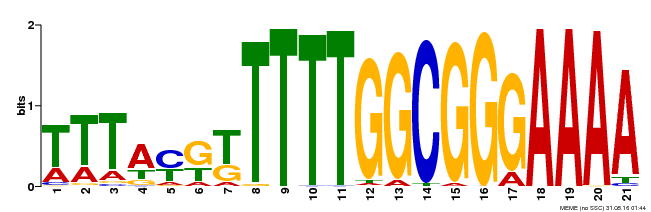
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK241993 | 0.0 | AK241993.1 Oryza sativa Japonica Group cDNA, clone: J075099J10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025878051.1 | 0.0 | B3 domain-containing protein LOC_Os12g40080 | ||||
| Swissprot | Q2QMT6 | 0.0 | Y1208_ORYSJ; B3 domain-containing protein LOC_Os12g40080 | ||||
| TrEMBL | A0A0E0FDM4 | 0.0 | A0A0E0FDM4_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI12G12100.1 | 0.0 | (Oryza meridionalis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G33280.1 | 6e-27 | B3 family protein | ||||



