 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA01G06510.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 366aa MW: 39043 Da PI: 9.5223 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 41.1 | 4.2e-13 | 109 | 153 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ ++ + ++lG+g+W+ I+r + Rt+ q+ s+ qky
ONIVA01G06510.1 109 PWTEEEHRMFLMGLQKLGKGDWRGISRNFVVSRTPTQVASHAQKY 153
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.60.10 | 0.001 | 2 | 23 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS50158 | 8.565 | 3 | 19 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 19.1 | 102 | 158 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.12E-16 | 104 | 158 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.0E-19 | 105 | 157 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-12 | 106 | 152 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.6E-11 | 106 | 156 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.83E-9 | 109 | 154 | No hit | No description |
| Pfam | PF00249 | 1.1E-10 | 109 | 153 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000122 | Biological Process | negative regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0030307 | Biological Process | positive regulation of cell growth | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:2000469 | Biological Process | negative regulation of peroxidase activity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
MTRRCSHCSN NGHNARTCPA RGGGGGGGGV RLFGVRLTSP PEVAMKKSAS MSCIASSLGS 60 GGGSGGSSPA GTGRGGGGGG EGAAGYASDD PTHASCSTNG RGERKKGTPW TEEEHRMFLM 120 GLQKLGKGDW RGISRNFVVS RTPTQVASHA QKYFIRQTNS SRRKRRSSLF DMVPEMPMDE 180 SPVVVEQLML HSTQDEATSS NQLPISHLVK QKEPEFARHL SDLQLRKHEE SEFTEPSLAA 240 LDLEMNHAAP FKTKFVLTMP TFYPALIPVP LTLWPPNVAN VGESGTNHEI LKPTPVNGKE 300 VINKADEVVG MSKLTIGDGS SNSIEPSALS LQLTGPTNTR QSAFHVNPPM AGPDLNKRNN 360 SPIHAV |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00551 | DAP | Transfer from AT5G47390 | Download |
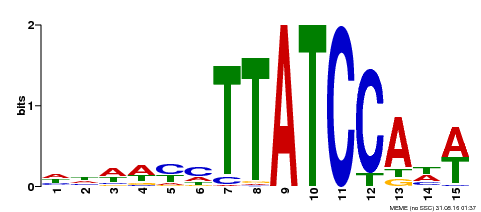
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK071611 | 0.0 | AK071611.1 Oryza sativa Japonica Group cDNA clone:J023105I20, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015621787.1 | 0.0 | transcription factor MYBS3-like | ||||
| TrEMBL | A0A0E0FHD2 | 0.0 | A0A0E0FHD2_ORYNI; Uncharacterized protein | ||||
| TrEMBL | A2WLJ1 | 0.0 | A2WLJ1_ORYSI; Uncharacterized protein | ||||
| TrEMBL | Q5SND9 | 0.0 | Q5SND9_ORYSJ; Os01g0187900 protein | ||||
| STRING | OS01T0187900-01 | 0.0 | (Oryza sativa) | ||||
| STRING | ONIVA01G06510.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1939 | 37 | 91 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47390.1 | 2e-75 | MYB_related family protein | ||||



