 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA01G09270.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 578aa MW: 60011.5 Da PI: 7.322 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 111.9 | 8.7e-35 | 211 | 311 | 2 | 86 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgt...............ssssaseceaessss 80
+g+kdrhsk++T++g+RdRRvRlsa++a++f+d+qd+LG+d++sk+++WL+++ak ai++l + ++++s++ ++s ++
ONIVA01G09270.1 211 TGRKDRHSKVCTARGPRDRRVRLSAHTAIQFYDVQDRLGYDRPSKAVDWLIKNAKDAIDKLDVLpawqptaggagagnaAAPPSSSTHPDSAEN 304
79**************************************************************888888888888776555555553333333 PP
TCP 81 sas.nss 86
s++ ++
ONIVA01G09270.1 305 SDDqAQA 311
3330222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 2.1E-30 | 211 | 284 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 33.257 | 213 | 271 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 578 aa Download sequence Send to blast |
METRPPAAPA KLSYGIRRGG WTRIGAATVA AGKKAAGDLD PRHHHHRVTH GGDGGGVGGG 60 GSGGQEEADE QQQQQHDHHR LLQLHHHQGV QQDQEPPPVP VFHLQPASVR QLSGSSAEYA 120 LLSPMGDAGG HSHHHQHGFQ PQLLSFGGVG HHHHLHQFTA QPQPPAASHT RGRGGGGEIV 180 PATTTPRSRG GGGGGGGEIV AVQGGHIVRS TGRKDRHSKV CTARGPRDRR VRLSAHTAIQ 240 FYDVQDRLGY DRPSKAVDWL IKNAKDAIDK LDVLPAWQPT AGGAGAGNAA APPSSSTHPD 300 SAENSDDQAQ AITVAHTAFD FAGGGSGGTS FLPPSLDSDA IADTIKSFFP MGGTAGGEAS 360 SSTTAAQSSA MGFQSYTPDL LSRTGSQSQE LRLSLQSLPD PMFHHQQHRH GGGGNGTTQQ 420 ALFSGAANYS FGGGAMWATE QQAQNQRMLP WNVPDPGGGG GAAYLFNVSQ QAAHMQAAAA 480 LGGHQSQFFF QRGPLQSSNQ PSERGWPETV EADNQMSHHQ GGLSPSVSAA IGFAAPGIGF 540 SGFRLPARIQ GDEEHNGGGG GNGDKPPPPS SVSSASHH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 1e-20 | 218 | 271 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 1e-20 | 218 | 271 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
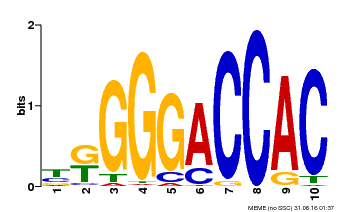
| UniProt | Transcription activator. Binds the promoter core sequence 5'-GGNCC-3'. {ECO:0000269|PubMed:12000681}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00009 | PBM | Transfer from 929286 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP003104 | 0.0 | AP003104.2 Oryza sativa Japonica Group genomic DNA, chromosome 1, BAC clone:OSJNBa0038J17. | |||
| GenBank | AP014957 | 0.0 | AP014957.1 Oryza sativa Japonica Group DNA, chromosome 1, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015634261.1 | 0.0 | transcription factor PCF5 | ||||
| Swissprot | A2WM14 | 0.0 | PCF5_ORYSI; Transcription factor PCF5 | ||||
| TrEMBL | A0A0E0FIF1 | 0.0 | A0A0E0FIF1_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA01G09270.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5839 | 33 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15030.3 | 7e-59 | TCP family protein | ||||



