 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA01G48030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 166aa MW: 18154.4 Da PI: 5.3046 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.5 | 7.9e-19 | 21 | 70 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++GVr ++ +g+++AeIrd + ng r++lg+f++aeeAa a+++a+ +++g
ONIVA01G48030.1 21 FRGVRKRP-WGKFAAEIRDSTRNG--VRVWLGTFDSAEEAALAYDQAAFAMRG 70
79******.**********44465..*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 22.682 | 20 | 78 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-30 | 20 | 79 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-36 | 20 | 84 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.3E-10 | 21 | 32 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.24E-22 | 21 | 80 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.38E-30 | 21 | 78 | No hit | No description |
| Pfam | PF00847 | 1.0E-13 | 21 | 70 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.3E-10 | 44 | 60 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 166 aa Download sequence Send to blast |
MDAHGQEEEE EPMQVQQQQA FRGVRKRPWG KFAAEIRDST RNGVRVWLGT FDSAEEAALA 60 YDQAAFAMRG SAAVLNFPME QVRRSMDMSL LQEGASPVVA LKRRHSMRAA AAGRRRKSAA 120 PAPADQEGGG GVMELEDLGP DYLEELLAAS QPIDITCCTS PSHHSI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-28 | 21 | 80 | 15 | 74 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 101 | 116 | KRRHSMRAAAAGRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May act as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. Involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250, ECO:0000269|PubMed:11950980}. | |||||
| UniProt | Transcription activator that binds to the GCC-box cis-acting elements found in the promoter regions of ethylene-responsive genes (PubMed:23057995). Acts downstream of MYC2 in the jasmonate-mediated response to Botrytis cinerea infection (PubMed:28733419). With MYC2 forms a transcription module that regulates pathogen-responsive genes (PubMed:28733419). {ECO:0000269|PubMed:23057995, ECO:0000269|PubMed:28733419}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00286 | DAP | Transfer from AT2G31230 | Download |
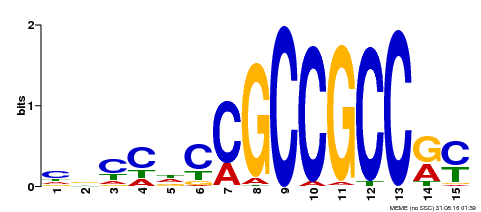
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding and infection with the fungal pathogen Botrytis cinerea. {ECO:0000269|PubMed:28733419}. | |||||
| UniProt | INDUCTION: Weakly induced by Pseudomonas syringae tomato (avirulent avrRpt2 strain), but also by mock inoculation. {ECO:0000269|PubMed:11950980}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC092263 | 0.0 | AC092263.7 Oryza sativa chromosome 3 BAC OSJNBa0033P04 genomic sequence, complete sequence. | |||
| GenBank | AK109390 | 0.0 | AK109390.2 Oryza sativa Japonica Group cDNA clone:006-211-G07, full insert sequence. | |||
| GenBank | AP014959 | 0.0 | AP014959.1 Oryza sativa Japonica Group DNA, chromosome 3, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629379.1 | 1e-119 | ethylene-responsive transcription factor 15 | ||||
| Swissprot | A0A3Q7I5Y9 | 5e-50 | ERFC3_SOLLC; Ethylene-response factor C3 | ||||
| Swissprot | Q8VYM0 | 1e-49 | ERF93_ARATH; Ethylene-responsive transcription factor 15 | ||||
| TrEMBL | A0A0D9ZG56 | 1e-118 | A0A0D9ZG56_9ORYZ; Uncharacterized protein | ||||
| TrEMBL | A0A0E0FY19 | 1e-118 | A0A0E0FY19_ORYNI; Uncharacterized protein | ||||
| TrEMBL | B8AP94 | 1e-118 | B8AP94_ORYSI; Uncharacterized protein | ||||
| TrEMBL | Q7Y1E5 | 1e-118 | Q7Y1E5_ORYSJ; Putative DNA binding protein | ||||
| STRING | OGLUM03G41640.1 | 1e-119 | (Oryza glumipatula) | ||||
| STRING | ONIVA01G48030.1 | 1e-119 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP277 | 37 | 249 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23240.1 | 2e-31 | ethylene response factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



