 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA02G02260.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 708aa MW: 79159.9 Da PI: 7.387 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 170.8 | 7.2e-53 | 57 | 208 | 1 | 142 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl..eeaeaagssasaspes 92
gg +++++++E+E++k+RER+RRai++++++GLR++Gn++lp+raD+n+Vl+AL+r+AGw+v++DGtt+r++s+pl ++++ g ++ +s+e+
ONIVA02G02260.1 57 GGEREREREREKERTKLRERHRRAITSRMLSGLRQHGNFPLPARADMNDVLAALARAAGWTVHPDGTTFRASSQPLhpPTPQSPGIFHVNSVET 150
4678899********************************************************************998999************* PP
DUF822 93 slq.sslkssalaspvesysaspksssfpspssldsislasa.......asllpvlsv 142
++ s+l+s+a+ +p++s+++++++++++spsslds+ +a++ ++ +v+s
ONIVA02G02260.1 151 PSFtSVLNSYAIGTPLDSQASMLQTDDSLSPSSLDSVVVADQsiknekyGNSDSVSSL 208
*99899*********************************9987766655444444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.3E-49 | 59 | 211 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 1.3E-158 | 229 | 662 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 3.91E-163 | 229 | 701 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 1.5E-69 | 254 | 610 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 267 | 281 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 288 | 306 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 401 | 423 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 500 | 516 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 517 | 528 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 535 | 558 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 8.4E-41 | 573 | 595 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 708 aa Download sequence Send to blast |
MSLKHPHSPV LDGDPPPHRR PRGLVSTPPP PAVAADPSPS PSPAAPPPRR RGGGGGGGER 60 EREREREKER TKLRERHRRA ITSRMLSGLR QHGNFPLPAR ADMNDVLAAL ARAAGWTVHP 120 DGTTFRASSQ PLHPPTPQSP GIFHVNSVET PSFTSVLNSY AIGTPLDSQA SMLQTDDSLS 180 PSSLDSVVVA DQSIKNEKYG NSDSVSSLNC LENHQLTRAS AALAGDYTRT PYIPVYASLP 240 MGIINSHCQL IDPEGIRAEL MHLKSLNVDG VIVDCWWGIV EAWIPHKYEW SGYRDLFGII 300 KEFKLKVQVV LSFHGSGETG SGGVSLPKWV MEIAQENQDV FFTDREGRRN MECLSWGIDK 360 ERVLRGRTGI EAYFDFMRSF HMEFRNLTEE GLISAIEIGL GVSGELKYPS CPERMGWRYP 420 GIVLRQNLRQ AALSRGHLFW ARGPDNAGYY NSRPHETGFF CDGGDYDSYY GRFFLNWYSG 480 ILIDHVDQVL SLATLAFDGV ETVVKIPSIY WWYRTASHAA ELTAGFYNPT NRDGYSPVFR 540 MLKKHSVILK FVCYGPEFTI QENNEAFADP EGLTWQVMNA AWDHGLSISV ESALPCLDGE 600 MYSQILDTAK PRHDPDRHHV SFFAYRQLPS FLLQRDVCFS ELGNFVKCMH DGSLIKIVVR 660 VRIIIAHQSG SSALDVLMQR LTVLCIRGVS TYPIKIIRSR YLTTVIAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-112 | 232 | 650 | 11 | 442 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
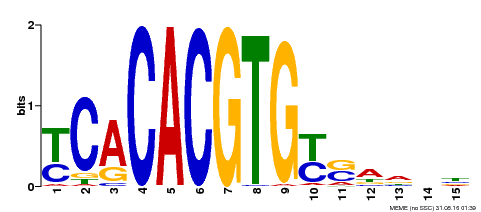
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK120049 | 0.0 | AK120049.1 Oryza sativa Japonica Group cDNA clone:J013000A07, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015622759.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A0E0G0K6 | 0.0 | A0A0E0G0K6_ORYNI; Beta-amylase | ||||
| STRING | ONIVA02G02260.2 | 0.0 | (Oryza nivara) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



