 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA02G22590.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 679aa MW: 74818.1 Da PI: 6.3001 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.4 | 2.7e-23 | 129 | 229 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+lt sd++++g +++ +++aee+ ++++ + +l+ +d++g++W +++i+r++++r++lt+GW+ Fv++++L +gD ++F +
ONIVA02G22590.1 129 FCKTLTASDTSTHGGFSVLRRHAEEClppldmtQNPPWQ--ELVARDLHGNEWHFRHIFRGQPRRHLLTTGWSVFVSSKRLVAGDAFIFL--RG 218
99****************************986555545..8************************************************..44 PP
SEE..EEEEE- CS
B3 88 sefelvvkvfr 98
+++el+v+v+r
ONIVA02G22590.1 219 ENGELRVGVRR 229
9999*****97 PP
| |||||||
| 2 | Auxin_resp | 111.4 | 9e-37 | 255 | 337 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+st++ F+v+Y+Pr+s+seFvv+++k+++a ++k+svGmRfkm+fe+++++err+sGt++gv ++++ W nS WrsLk
ONIVA02G22590.1 255 ASHAISTGTLFSVFYKPRTSQSEFVVSANKYLEAKNSKISVGMRFKMRFEGDEAPERRFSGTIIGVGSMSTSPWANSDWRSLK 337
789******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 4.58E-44 | 118 | 258 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 4.6E-41 | 124 | 244 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.78E-22 | 127 | 229 | No hit | No description |
| SMART | SM01019 | 1.5E-22 | 129 | 231 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.462 | 129 | 231 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.4E-21 | 129 | 229 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.9E-34 | 255 | 337 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 5.1E-13 | 536 | 640 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.456 | 549 | 642 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 6.54E-12 | 551 | 628 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 679 aa Download sequence Send to blast |
MAGSVVAAAA AAGGGGTGSS CDALYRELWH ACAGPLVTVP RQGELVYYFP QGHMEQLEAS 60 TDQQLDQHLP LFNLPSKILC KVVNVELRAE TDSDEVYAQI MLQPEADQNE LTSPKPEPHE 120 PEKCNVHSFC KTLTASDTST HGGFSVLRRH AEECLPPLDM TQNPPWQELV ARDLHGNEWH 180 FRHIFRGQPR RHLLTTGWSV FVSSKRLVAG DAFIFLRGEN GELRVGVRRL MRQLNNMPSS 240 VISSHSMHLG VLATASHAIS TGTLFSVFYK PRTSQSEFVV SANKYLEAKN SKISVGMRFK 300 MRFEGDEAPE RRFSGTIIGV GSMSTSPWAN SDWRSLKVQW DEPSVVPRPD RVSPWELEPL 360 AVSNSQPSPQ PPARNKRARP PASSSIAPEL PPVFGLWKSS AESTQGFSFS GLQRTQELYP 420 SSPNPIFSTS LNVGFSTKNE PSALSNKHFY WPMRETRADS YSASISKVPS EKKQEPSSAG 480 CRLFGIEISS AVEATSPLAA VSGVGQDQPA ASVDAESDQL SQPSHANKSD APAASSEPSP 540 HETQSRQVRS CTKVIMQGMA VGRAVDLTRL HGYDDLRCKL EEMFDIQGEL SASLKKWKVV 600 YTDDEDDMML VGDDPWPEFC SMVKRIYIYT YEEAKQLTPK SKLPIIGDAI KPNPNKQSPE 660 SDMPHSDLDS TAPVTDKDC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 0.0 | 6 | 360 | 1 | 355 | Auxin response factor 1 |
| 4ldw_A | 0.0 | 6 | 360 | 1 | 355 | Auxin response factor 1 |
| 4ldw_B | 0.0 | 6 | 360 | 1 | 355 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00097 | PBM | Transfer from AT1G59750 | Download |
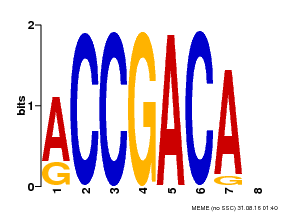
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK070026 | 0.0 | AK070026.1 Oryza sativa Japonica Group cDNA clone:J023036G16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625501.1 | 0.0 | auxin response factor 7 | ||||
| Swissprot | Q6YVY0 | 0.0 | ARFG_ORYSJ; Auxin response factor 7 | ||||
| TrEMBL | A0A0E0G882 | 0.0 | A0A0E0G882_ORYNI; Auxin response factor | ||||
| STRING | ONIVA02G22590.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1082 | 36 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59750.4 | 0.0 | auxin response factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



