 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA04G14730.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 849aa MW: 91897.4 Da PI: 9.131 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 83.5 | 2.8e-26 | 513 | 581 | 2 | 70 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekk 70
W ++e+l+Li++rrem+++++++k++k+lWe++s++mre+gf rsp++C++kw+nl k++kk + ++++
ONIVA04G14730.1 513 WVQDETLCLIALRREMDSHFNTSKSNKHLWEAISARMREQGFDRSPTMCTDKWRNLLKEFKKARSHARG 581
***************************************************************999876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81383 | 1.8E-5 | 19 | 69 | IPR001810 | F-box domain |
| PROSITE profile | PS50090 | 8.293 | 505 | 569 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-4 | 506 | 573 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-5 | 509 | 571 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 3.78E-29 | 511 | 576 | No hit | No description |
| Pfam | PF13837 | 1.3E-17 | 512 | 583 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 849 aa Download sequence Send to blast |
MSSSSSSPRA PGDRKRCRRA SGPVPRWASL PEDLVDLVAS RLLAGGDLLD YVRFRAVCTS 60 WRSGTASPRG RGVADRRFHP RRWMMLPEGH GLYPGHPSLR GYARFLNLDT GTLVRARIPL 120 LRDGYVAIDS VDGLLLLLLD PDPNQEGAVR LLRPFTGDTA ELPPLGTVLP HLGSRLLDCP 180 APATGAGAGA ITVLLALSVV SRVAFATSLD RQWSLSTYEC VTLSSPIASH GKIYLMHTDR 240 SCGEKMHQIL RIDHPPAAAQ DGSGSGAGRA LQEPKLVATI PARKLDHFQG LVECGSEILV 300 LGYKNWSTSR ISVFKLADLV LQRFMPIKSI EGHTLFIGER NISVSSKILP TVKGDNLVYL 360 NSGLVKYHLS SGSLSLAIDN CSLYGRAPGP SSLVHYIYSC CIRNRWELGP GRAERPSHLP 420 RVPPGSPRLR SPTAATTAPV TSPIAGFPFA HGTTPRAPPR PAAAMLLSGP SPQQPTPPLL 480 LPESSGEDGG HDSSSRAAAS GGGGGPKKRA ETWVQDETLC LIALRREMDS HFNTSKSNKH 540 LWEAISARMR EQGFDRSPTM CTDKWRNLLK EFKKARSHAR GGGGGGVGGG GAGTGGGNCP 600 AKMACYKEID DLLKRRGKPT GGGGAAVGSG AVKSPTVTSK IDSYLQFDKG FEDASIPFGP 660 VEASGRSLLS VEDRLEPDSH PLALTADAVA TNGVNPWNWR DTSTNGGDNQ VTFGGRVILV 720 KWGDYTKRIG IDGTAEAIKE AIKSAFGLRT RRAFWLEDED EVVRSLDRDM PVGTYTLHLD 780 TGMTIKLYMF ENDEVRTEDK TFYTEEDFRD FLSRRGWTLL REYSGYRIAD TLDDLRPGVI 840 YEGMRSLGD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 4e-38 | 507 | 608 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00640 | PBM | Transfer from MDP0000164819 | Download |
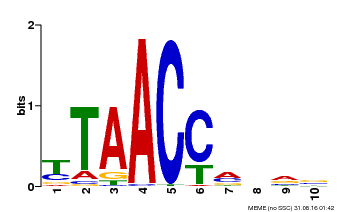
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK103296 | 0.0 | AK103296.1 Oryza sativa Japonica Group cDNA clone:J033125B02, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0E0H2B9 | 0.0 | A0A0E0H2B9_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA04G14730.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5631 | 37 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 1e-128 | Trihelix family protein | ||||



