 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA04G15310.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 523aa MW: 55501.8 Da PI: 4.9905 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.7 | 9.8e-17 | 56 | 103 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WTt Ed lv+ v+q+G g+W+++ r+ g+ R++k+c++rw ++l
ONIVA04G15310.1 56 KGPWTTAEDAVLVQHVRQHGEGNWNAVQRMTGLLRCGKSCRLRWTNHL 103
79******************************99***********996 PP
| |||||||
| 2 | Myb_DNA-binding | 50.9 | 3.7e-16 | 109 | 152 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g+++++E++l++++++qlG++ W++ a++++ gRt++++k++w++
ONIVA04G15310.1 109 KGSFSPDEELLIAQLHAQLGNK-WARMASHLP-GRTDNEIKNYWNT 152
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.804 | 51 | 103 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.2E-14 | 55 | 105 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.95E-28 | 55 | 150 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.1E-14 | 56 | 103 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.3E-21 | 57 | 110 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.64E-11 | 58 | 103 | No hit | No description |
| PROSITE profile | PS51294 | 22.55 | 104 | 158 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.0E-15 | 108 | 156 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.2E-15 | 109 | 152 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.87E-11 | 111 | 152 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-24 | 111 | 157 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 523 aa Download sequence Send to blast |
MEEMIKPALA PASSEEEKEV VATAATGERR HEEATAGREE QEEEEEEEEA PVVLKKGPWT 60 TAEDAVLVQH VRQHGEGNWN AVQRMTGLLR CGKSCRLRWT NHLRPNLKKG SFSPDEELLI 120 AQLHAQLGNK WARMASHLPG RTDNEIKNYW NTRTKRRQRA GLPVYPPDVQ LHLAFAKRCR 180 YDDFSSPLSS PQQSAGSNVL SMDAADAAGA ASSGYTSARP PPLDLAGQLA MGSRPVQLLA 240 ATPFSAPSSP WGKPFARNAQ FFQFPHSSPV SPTTPTGPVQ VHPVTPELSL GYGLHAGDRA 300 RLPPVSPSPG ARAELPSSQL RPSMAPTTAA AAATGGLVGG ALQDHPNAAS LEAMLQELHD 360 AIKIEPPAPP ENRGTEEEGG GGGGNLRGFL HEQIFDKLMP IKSGDFCIVG LHSVLISLQF 420 PGDGKPEVEL KDDIETLFDL IIPATFPAAA PEPAAAATAA SAAPNHSSSV SQHSSDDQDH 480 SNGADVVLDL PILTGGGGGS SEQDDWSLDG AACQWDNISG GIC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 7e-29 | 54 | 157 | 25 | 127 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
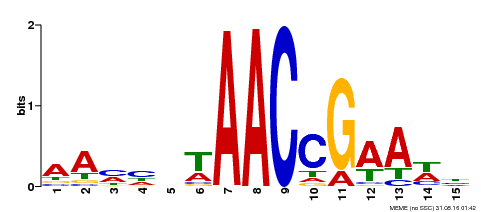
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012611 | 0.0 | CP012611.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 3 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629365.1 | 0.0 | transcription factor MYB65 | ||||
| TrEMBL | A0A0E0H2J7 | 0.0 | A0A0E0H2J7_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA04G15310.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7420 | 37 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.2 | 8e-66 | myb domain protein 101 | ||||



