 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA04G26660.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 415aa MW: 44907.5 Da PI: 7.8038 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.6 | 2.3e-40 | 183 | 261 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EE.ETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvl.vsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cq+egC+adls+ak+yhrrhkvCe+hskapvv+ + gl+qrfCqqCsrfh l+efD++k+sCr+rLa+hn+rrrk+++
ONIVA04G26660.1 183 RCQAEGCKADLSSAKRYHRRHKVCEHHSKAPVVVtAGGLHQRFCQQCSRFHLLDEFDDAKKSCRKRLADHNRRRRKSKP 261
6******************************98725689*************************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.0E-33 | 178 | 246 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 30.291 | 181 | 259 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 6.8E-37 | 182 | 262 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 8.6E-31 | 184 | 258 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MMNVPSAAAA SSCDDFGYNA TPPPPPSLLP IMDQDGGGGN IQRDHHHHHH QQLGYNLEPS 60 SLALLPPSNA AAAAAHHATI AHASPHDLLQ FYPTSHYLAA AGGAGGGGNP YSHFTAAAAA 120 GSTFQSYYQQ PPQAAPEYYF PTLVSSAEEN MASFAATQLG LNLGYRTYFP PRGGYTYGHH 180 PPRCQAEGCK ADLSSAKRYH RRHKVCEHHS KAPVVVTAGG LHQRFCQQCS RFHLLDEFDD 240 AKKSCRKRLA DHNRRRRKSK PSDGEHSGEK RRAQANKSAA TKDKAGSSSK NAGIGDGFET 300 QLLGGAHMSK DQDQAMDLGE VVKEAVDPKG KASMQQQQQQ AHHGIHQQSH QQHGFPFPSS 360 SGSCLFPQSQ GAVSSTDTSN IAQVQEPSLA FHQQHHQHSN ILQLGQAMFD LDFDH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-27 | 184 | 258 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 253 | 271 | RRRRKSKPSDGEHSGEKRR |
| 2 | 254 | 259 | RRRKSK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that plays an important role in building the laminar joint between leaf blade and leaf sheath boundary, thereby controlling ligule and auricle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
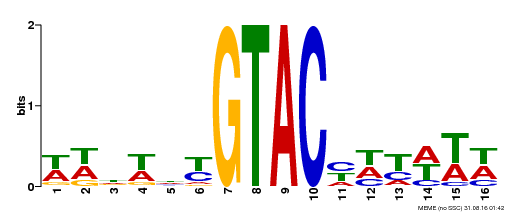
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB776992 | 0.0 | AB776992.1 Oryza rufipogon LG1 mRNA for squamosa promoter-binding-like protein 8, complete cds, strain: W630. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015634037.1 | 0.0 | squamosa promoter-binding-like protein 8 | ||||
| Swissprot | Q01KM7 | 0.0 | SPL8_ORYSI; Squamosa promoter-binding-like protein 8 | ||||
| TrEMBL | A0A0E0H6X9 | 0.0 | A0A0E0H6X9_ORYNI; Uncharacterized protein | ||||
| TrEMBL | L8B8J8 | 0.0 | L8B8J8_ORYRU; Squamosa promoter-binding-like protein 8 | ||||
| STRING | ONIVA04G26660.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6990 | 34 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.1 | 3e-42 | squamosa promoter binding protein-like 8 | ||||



