 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA05G01580.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 438aa MW: 49872.7 Da PI: 8.5836 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 20.1 | 1.7e-06 | 80 | 102 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cg sF+ + +L++H+++H
ONIVA05G01580.1 80 HDCKVCGASFKKPAHLRQHMQSH 102
56*******************99 PP
| |||||||
| 2 | zf-C2H2 | 19.5 | 2.7e-06 | 166 | 190 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ C+ s+srk++L+rH+ tH
ONIVA05G01580.1 166 FSCHvdGCPFSYSRKDHLNRHLLTH 190
89*********************99 PP
| |||||||
| 3 | zf-C2H2 | 11 | 0.0013 | 195 | 220 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+ Cp C++ F+ k n +rH + H
ONIVA05G01580.1 195 FACPmeGCNRKFTIKGNIQRHVQEmH 220
78*******************98877 PP
| |||||||
| 4 | zf-C2H2 | 20.5 | 1.2e-06 | 232 | 256 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp +Cgk+F+ s L++H +H
ONIVA05G01580.1 232 FICPeeNCGKTFKYASKLQKHEESH 256
78*******************9988 PP
| |||||||
| 5 | zf-C2H2 | 16.7 | 2.1e-05 | 323 | 347 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
+C+ dC+ sFs ksnL +H + H
ONIVA05G01580.1 323 RCHlkDCKLSFSKKSNLDKHVKAvH 347
7*******************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 6.78E-7 | 31 | 102 | No hit | No description |
| SMART | SM00355 | 15 | 33 | 55 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.385 | 80 | 107 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.7E-5 | 80 | 103 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0016 | 80 | 102 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 82 | 102 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.2E-9 | 163 | 192 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.88E-6 | 163 | 192 | No hit | No description |
| PROSITE profile | PS50157 | 9.349 | 166 | 195 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0016 | 166 | 190 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 168 | 190 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-6 | 193 | 221 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.18E-6 | 193 | 221 | No hit | No description |
| PROSITE profile | PS50157 | 10.866 | 195 | 225 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0019 | 195 | 220 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 197 | 220 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-7 | 228 | 256 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.57E-5 | 229 | 258 | No hit | No description |
| PROSITE profile | PS50157 | 11.24 | 232 | 256 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0018 | 232 | 256 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 234 | 256 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 7.8 | 264 | 289 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 266 | 289 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 13 | 292 | 313 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 3.19E-7 | 303 | 364 | No hit | No description |
| PROSITE profile | PS50157 | 11.967 | 322 | 352 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.031 | 322 | 347 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-5 | 323 | 348 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 324 | 347 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 7.2E-6 | 349 | 375 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 8.974 | 353 | 384 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 3.5 | 353 | 379 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 355 | 379 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 438 aa Download sequence Send to blast |
MGSVELGAEE REVAGGEGGS KGAAPPARDI RRYKCDFCSV VRSKKGLIRA HVLEHHKDEV 60 DDLDDYLGRG GGETCKEMDH DCKVCGASFK KPAHLRQHMQ SHSLEHWIPE NSKMREGLEW 120 TSTRMGRPFW EAPSRMPLSR SSSGWGDSPE KLHQECLVHT LFQRPFSCHV DGCPFSYSRK 180 DHLNRHLLTH QGKLFACPME GCNRKFTIKG NIQRHVQEMH KDGSPCESKK EFICPEENCG 240 KTFKYASKLQ KHEESHVKLD YSEVICCEPG CMKAFTNLEC LKAHNKSCHR HVVCDVCGTK 300 QLKKNFKRHQ RMHEGSCVTE RVRCHLKDCK LSFSKKSNLD KHVKAVHEQK RPFVCGFSGC 360 GKSFSYKHVR DNHEKSSAHV YVQANFEEID GERPRQAGGR KRKAIPVESL MRKRVAAPDD 420 DAPACDDGTE YLRWLLSG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 2e-13 | 163 | 286 | 40 | 157 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 2e-13 | 163 | 286 | 40 | 157 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
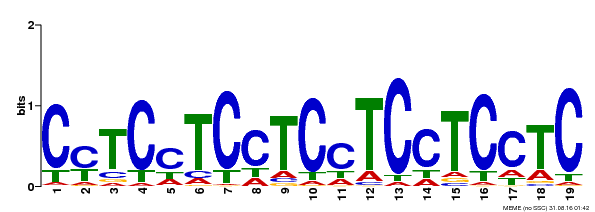
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK101631 | 0.0 | AK101631.1 Oryza sativa Japonica Group cDNA clone:J033054O17, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015637660.1 | 0.0 | transcription factor IIIA isoform X1 | ||||
| TrEMBL | A0A0E0H8S8 | 0.0 | A0A0E0H8S8_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA05G01580.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-90 | transcription factor IIIA | ||||



