 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA05G26000.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 394aa MW: 41376.9 Da PI: 10.055 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 39.5 | 1.4e-12 | 67 | 115 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ r +r++lg+f + eAa+a++ a+++++g
ONIVA05G26000.1 67 SKYKGVVPQP-NGRWGAQIYE------RhQRVWLGTFTGEAEAARAYDVAAQRFRG 115
89****9888.8*********......44**********99*************98 PP
| |||||||
| 2 | B3 | 93.3 | 1.7e-29 | 193 | 306 | 1 | 92 |
EEEE-..-HHHHTT-EE--HHH.HTT......................---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHH CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......................ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkan 72
f+k+ tpsdv+k++rlv+pk++ae+h gg + +++ l +ed g++W+++++y+++s++yvltkGW++Fvk++
ONIVA05G26000.1 193 FDKTVTPSDVGKLNRLVIPKQHAEKHfplqlppptttssvaaaadaaaGGGECKGVLLNFEDAAGKVWKFRYSYWNSSQSYVLTKGWSRFVKDK 286
89***************************99999999888888888885566699*************************************** PP
T--TT-EEEEEE-SSSEE.. CS
B3 73 gLkegDfvvFkldgrsefel 92
gL +gD v F++ ++++l
ONIVA05G26000.1 287 GLHAGDAVGFYRAAGKNAQL 306
***********765555555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.7E-16 | 67 | 123 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.7E-8 | 67 | 115 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.98E-23 | 67 | 123 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 1.5E-19 | 68 | 123 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.2E-26 | 68 | 129 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.862 | 68 | 123 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 1.1E-39 | 183 | 320 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.83E-28 | 191 | 311 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.12E-26 | 191 | 298 | No hit | No description |
| Pfam | PF02362 | 1.2E-26 | 193 | 308 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 6.1E-21 | 193 | 313 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.167 | 193 | 314 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 394 aa Download sequence Send to blast |
MDSTSCLLDD ASSGASTGKK AAAAAASKAL QRVGSGASAV MDAAEPGAEA DSGGERRGGG 60 GGKLPSSKYK GVVPQPNGRW GAQIYERHQR VWLGTFTGEA EAARAYDVAA QRFRGRDAVT 120 NFRPLAESDP EAAVELRFLA SRSKAEVVDM LRKHTYLEEL TQNKRAFAAI SPPPPKHPAS 180 SPPSSSAARE HLFDKTVTPS DVGKLNRLVI PKQHAEKHFP LQLPPPTTTS SVAAAADAAA 240 GGGECKGVLL NFEDAAGKVW KFRYSYWNSS QSYVLTKGWS RFVKDKGLHA GDAVGFYRAA 300 GKNAQLFIDC KVRAKPTTAA AAAAFLSAVA AAAAPPPAVK AIRLFGVDLL TAAAPELQDA 360 GGAAMTKSKR AMDAMAESQA HVVFKKQCIE LALT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 1e-48 | 190 | 314 | 11 | 118 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
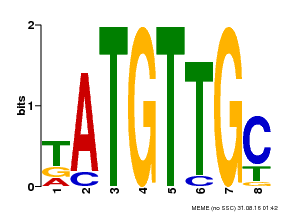
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012613 | 0.0 | CP012613.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 5 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015637817.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os05g0549800 | ||||
| Swissprot | Q6L4H4 | 0.0 | Y5498_ORYSJ; AP2/ERF and B3 domain-containing protein Os05g0549800 | ||||
| TrEMBL | A0A0E0HHS8 | 0.0 | A0A0E0HHS8_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA05G26000.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25560.1 | 6e-95 | RAV family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



