 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA07G22160.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1028aa MW: 114431 Da PI: 5.6683 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 180.4 | 2.1e-56 | 21 | 137 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+ ++rwl+++ei++iL n++k++++ e+++rp sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+v+vl+cyYah+een++fq
ONIVA07G22160.1 21 LEAQNRWLRPTEICHILSNYKKFSIAPEPPNRPASGSLFLFDRKILRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSVDVLHCYYAHGEENENFQ 114
5679****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rr+ywlLee + +ivlvhylevk
ONIVA07G22160.1 115 RRTYWLLEEGFMNIVLVHYLEVK 137
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.91 | 16 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.5E-76 | 19 | 137 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.4E-50 | 22 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.6E-5 | 432 | 519 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.71E-16 | 433 | 519 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.5E-4 | 433 | 506 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 1.71E-16 | 620 | 726 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-17 | 625 | 727 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.10E-13 | 626 | 724 | No hit | No description |
| PROSITE profile | PS50297 | 17.263 | 632 | 726 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.3E-7 | 638 | 727 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.618 | 665 | 697 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.055 | 665 | 694 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 670 | 704 | 733 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 4.2 | 838 | 860 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.718 | 839 | 868 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 2.1E-7 | 840 | 889 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.0055 | 840 | 859 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0096 | 861 | 883 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 862 | 886 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.4E-4 | 864 | 883 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1028 aa Download sequence Send to blast |
MAEVRKYGLP NQPPDIPQIL LEAQNRWLRP TEICHILSNY KKFSIAPEPP NRPASGSLFL 60 FDRKILRYFR KDGHNWRKKK DGKTVKEAHE KLKVGSVDVL HCYYAHGEEN ENFQRRTYWL 120 LEEGFMNIVL VHYLEVKGGK QSFSRSKEAE ESAGLSNADS PACSNSFASQ SQVASQSMDA 180 ESPISGQISE YEDAETDNCR ASSRYHPFVE MQQPVDGVMM NNMLGVSAPS TSTTTANSDN 240 HFATHYDIAG VFNEAGAGLR GVSKTLHDSV RFAEPYPECS AEFMEPALYS SNATMESNNL 300 DDNSRLETFM SEALYTNNLT QKEADALSAA GIMSSQAENN SYTDGIRYPL LKQSSLDLFK 360 IEPDGLKKFD SFSRWMSSEL PEVADLDIKS SSDAFWSSTE TVNVADGTSI PINEQLDAFA 420 VSPSLSQDQL FSIIDVSPSY ACTGSRNKVL ITGTFLANKE HVENCKWSCM FGDVEVPAEV 480 LAHGSLRCYT PVHLSGRVPF YVTCSNRVAC SEVREFEFRD SDARQMDTSD PQTTGINEMH 540 LHIRLEKLLS LGPDDYEKYV MSDGKEKSEI INTISSLMLD DKCLNQAVPL DEKEVSTARD 600 QNIEKLVKEK LYCWLVHKVH DEDKGPNVLG KEGQGVIHLV AALGYDWAVR PIITAGVKVN 660 FRDARGWTAL HWAASCGRER TVGALIANGA ESGLLTDPTP QFPAGRTAAD LASENGHKGI 720 AGFLAESALT SHLSALTLKE SKDGNVKEIC GLGGAEDFAE SSSAQLAYRD SQAESLKDSL 780 SAVRKSTQAA ARIFQAFRVE SFHRKKVVEY GDDDCGLSDE RTLSLVSIKN AKPGQNDGSH 840 SAAVRIQNKF RGWKGRKEFM IIRQKIVKIQ AHVRGHQVRK SYRRIVWSVG IVEKIILRWR 900 RKRRGLRGFQ PVKQLEGPSP IQQLEGPSQI QPAKEEEEDE YDYLKDGRKQ AEGRLQRALA 960 RVKSMTQYPE AREQYSRIAN RVTELQEPQA MMIQDDMQSD GAIADGGDFM AELEELCGDG 1020 DAPMPTIL |
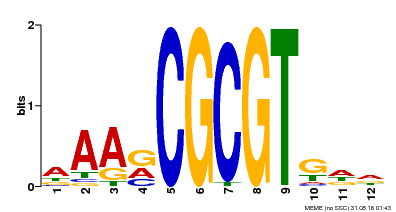
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK108461 | 0.0 | AK108461.1 Oryza sativa Japonica Group cDNA clone:002-143-D08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647012.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| TrEMBL | A0A0E0I479 | 0.0 | A0A0E0I479_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA07G22160.2 | 0.0 | (Oryza nivara) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||



