 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA07G22160.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1034aa MW: 115065 Da PI: 5.6702 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 180.4 | 2.2e-56 | 21 | 137 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+ ++rwl+++ei++iL n++k++++ e+++rp sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+v+vl+cyYah+een++fq
ONIVA07G22160.2 21 LEAQNRWLRPTEICHILSNYKKFSIAPEPPNRPASGSLFLFDRKILRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSVDVLHCYYAHGEENENFQ 114
5679****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rr+ywlLee + +ivlvhylevk
ONIVA07G22160.2 115 RRTYWLLEEGFMNIVLVHYLEVK 137
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.91 | 16 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.5E-76 | 19 | 137 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.4E-50 | 22 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.6E-5 | 438 | 525 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.71E-16 | 439 | 525 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.5E-4 | 439 | 512 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 1.71E-16 | 626 | 732 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-17 | 631 | 733 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.16E-13 | 632 | 730 | No hit | No description |
| PROSITE profile | PS50297 | 17.263 | 638 | 732 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.4E-7 | 644 | 733 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.618 | 671 | 703 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.055 | 671 | 700 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 670 | 710 | 739 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 4.2 | 844 | 866 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.718 | 845 | 874 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 2.12E-7 | 846 | 895 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.0056 | 846 | 865 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0096 | 867 | 889 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 868 | 892 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.4E-4 | 870 | 889 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1034 aa Download sequence Send to blast |
MAEVRKYGLP NQPPDIPQIL LEAQNRWLRP TEICHILSNY KKFSIAPEPP NRPASGSLFL 60 FDRKILRYFR KDGHNWRKKK DGKTVKEAHE KLKVGSVDVL HCYYAHGEEN ENFQRRTYWL 120 LEEGFMNIVL VHYLEVKGGK QSFSRSKEAE ESAGLSNADS PACSNSFASQ SQVASQSMDA 180 ESPISGQISE YEDAETEKFG ATDNCRASSR YHPFVEMQQP VDGVMMNNML GVSAPSTSTT 240 TANSDNHFAT HYDIAGVFNE AGAGLRGVSK TLHDSVRFAE PYPECSAEFM EPALYSSNAT 300 MESNNLDDNS RLETFMSEAL YTNNLTQKEA DALSAAGIMS SQAENNSYTD GIRYPLLKQS 360 SLDLFKIEPD GLKKFDSFSR WMSSELPEVA DLDIKSSSDA FWSSTETVNV ADGTSIPINE 420 QLDAFAVSPS LSQDQLFSII DVSPSYACTG SRNKVLITGT FLANKEHVEN CKWSCMFGDV 480 EVPAEVLAHG SLRCYTPVHL SGRVPFYVTC SNRVACSEVR EFEFRDSDAR QMDTSDPQTT 540 GINEMHLHIR LEKLLSLGPD DYEKYVMSDG KEKSEIINTI SSLMLDDKCL NQAVPLDEKE 600 VSTARDQNIE KLVKEKLYCW LVHKVHDEDK GPNVLGKEGQ GVIHLVAALG YDWAVRPIIT 660 AGVKVNFRDA RGWTALHWAA SCGRERTVGA LIANGAESGL LTDPTPQFPA GRTAADLASE 720 NGHKGIAGFL AESALTSHLS ALTLKESKDG NVKEICGLGG AEDFAESSSA QLAYRDSQAE 780 SLKDSLSAVR KSTQAAARIF QAFRVESFHR KKVVEYGDDD CGLSDERTLS LVSIKNAKPG 840 QNDGSHSAAV RIQNKFRGWK GRKEFMIIRQ KIVKIQAHVR GHQVRKSYRR IVWSVGIVEK 900 IILRWRRKRR GLRGFQPVKQ LEGPSPIQQL EGPSQIQPAK EEEEDEYDYL KDGRKQAEGR 960 LQRALARVKS MTQYPEAREQ YSRIANRVTE LQEPQAMMIQ DDMQSDGAIA DGGDFMAELE 1020 ELCGDGDAPM PTIL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
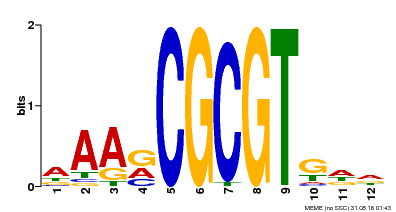
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK108461 | 0.0 | AK108461.1 Oryza sativa Japonica Group cDNA clone:002-143-D08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647012.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| TrEMBL | A0A0E0I480 | 0.0 | A0A0E0I480_ORYNI; Uncharacterized protein | ||||
| STRING | ONIVA07G22160.2 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP608 | 38 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||



