 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA07G22160.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 999aa MW: 111295 Da PI: 5.6659 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 180.5 | 2.1e-56 | 21 | 137 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+ ++rwl+++ei++iL n++k++++ e+++rp sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+v+vl+cyYah+een++fq
ONIVA07G22160.3 21 LEAQNRWLRPTEICHILSNYKKFSIAPEPPNRPASGSLFLFDRKILRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSVDVLHCYYAHGEENENFQ 114
5679****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rr+ywlLee + +ivlvhylevk
ONIVA07G22160.3 115 RRTYWLLEEGFMNIVLVHYLEVK 137
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.91 | 16 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.5E-76 | 19 | 137 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.2E-50 | 22 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.3E-5 | 403 | 490 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.71E-16 | 404 | 490 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.4E-4 | 404 | 477 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 1.71E-16 | 591 | 697 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.6E-17 | 596 | 698 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 6.82E-13 | 597 | 695 | No hit | No description |
| PROSITE profile | PS50297 | 17.263 | 603 | 697 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.2E-7 | 609 | 698 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.055 | 636 | 665 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.618 | 636 | 668 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 670 | 675 | 704 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 4.2 | 809 | 831 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.718 | 810 | 839 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 2.04E-7 | 811 | 860 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.0054 | 811 | 830 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0096 | 832 | 854 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 833 | 857 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.3E-4 | 835 | 854 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 999 aa Download sequence Send to blast |
MAEVRKYGLP NQPPDIPQIL LEAQNRWLRP TEICHILSNY KKFSIAPEPP NRPASGSLFL 60 FDRKILRYFR KDGHNWRKKK DGKTVKEAHE KLKVGSVDVL HCYYAHGEEN ENFQRRTYWL 120 LEEGFMNIVL VHYLEVKGGK QSFSRSKEAE ESAGLSNADS PACSNSFASQ SQVASQSMDA 180 ESPISGQISE YEDAETGYHG EMQTTTANSD NHFATHYDIA GVFNEAGAGL RGVSKTLHDS 240 VRFAEPYPEC SAEFMEPALY SSNATMESNN LDDNSRLETF MSEALYTNNL TQKEADALSA 300 AGIMSSQAEN NSYTDGIRYP LLKQSSLDLF KIEPDGLKKF DSFSRWMSSE LPEVADLDIK 360 SSSDAFWSST ETVNVADGTS IPINEQLDAF AVSPSLSQDQ LFSIIDVSPS YACTGSRNKV 420 LITGTFLANK EHVENCKWSC MFGDVEVPAE VLAHGSLRCY TPVHLSGRVP FYVTCSNRVA 480 CSEVREFEFR DSDARQMDTS DPQTTGINEM HLHIRLEKLL SLGPDDYEKY VMSDGKEKSE 540 IINTISSLML DDKCLNQAVP LDEKEVSTAR DQNIEKLVKE KLYCWLVHKV HDEDKGPNVL 600 GKEGQGVIHL VAALGYDWAV RPIITAGVKV NFRDARGWTA LHWAASCGRE RTVGALIANG 660 AESGLLTDPT PQFPAGRTAA DLASENGHKG IAGFLAESAL TSHLSALTLK ESKDGNVKEI 720 CGLGGAEDFA ESSSAQLAYR DSQAESLKDS LSAVRKSTQA AARIFQAFRV ESFHRKKVVE 780 YGDDDCGLSD ERTLSLVSIK NAKPGQNDGS HSAAVRIQNK FRGWKGRKEF MIIRQKIVKI 840 QAHVRGHQVR KSYRRIVWSV GIVEKIILRW RRKRRGLRGF QPVKQLEGPS PIQQLEGPSQ 900 IQPAKEEEED EYDYLKDGRK QAEGRLQRAL ARVKSMTQYP EAREQYSRIA NRVTELQEPQ 960 AMMIQDDMQS DGAIADGGDF MAELEELCGD GDAPMPTIL |
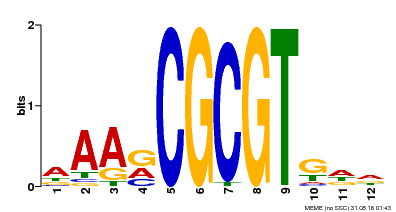
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK108461 | 0.0 | AK108461.1 Oryza sativa Japonica Group cDNA clone:002-143-D08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647012.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| TrEMBL | A0A0E0I481 | 0.0 | A0A0E0I481_ORYNI; Uncharacterized protein | ||||
| STRING | ORUFI07G23760.2 | 0.0 | (Oryza rufipogon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.1 | 0.0 | ethylene induced calmodulin binding protein | ||||



