 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ONIVA10G01080.4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 332aa MW: 36483.6 Da PI: 10.0996 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 382.8 | 5.4e-117 | 1 | 328 | 1 | 300 |
GAGA_bind 1 mdddgsre.rnkg.yyepaaslkenlglqlmssia.erdaki...........rernlalsekkaavaerd..................maflq 62
mddd+s++ r +g ++e+ ++nlglqlmss++ +rd+k+ +++++ +++++++++rd +
ONIVA10G01080.4 1 MDDDASMSiR-WGgFFESP---ARNLGLQLMSSVPaDRDTKQllsgspflhhqHQQQHVPHHHHQPHHPRDcgangnanggamppppatE---- 86
9*******66.777***99...88**********99***************99777777***********************99976431.... PP
GAGA_bind 63 rdkalaernkalverdnkllalllvensla...salpvgvqvlsgtksidslqqlsepqledsave.lreeeklealpieeaaeeakekkkkkk 152
+ + +++++r++k+l++l+v ++++ +++pvg+++++gt++++++qq++epq ++++ + +++ee+++++ iee++++++e + +kk
ONIVA10G01080.4 87 APPSMPMNFQHHHPREHKVLHNLTVGHGSShiaHHDPVGYGMIPGTHTLQMMQQQTEPQPQPPPPPqQPKEECISSPLIEENVPVIDEPPPPKK 180
333444444899***************9988999***************************99999899************************* PP
GAGA_bind 153 rqrakkpkekkakkkkkksekskkkvkkesad.erskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLP 245
rq++++pk+++akk+kk s++++++ ++++a+ +r++ ++k+i++v+ng++lD s++P+PvCsCtGa++qCY+WG+GGWqSaCCtttiS+yPLP
ONIVA10G01080.4 181 RQQGRQPKVPRAKKPKK-SAAPREDGAPPNAPaPRRRGPRKNIGMVINGIDLDLSRIPTPVCSCTGAPQQCYRWGAGGWQSACCTTTISTYPLP 273
*****************.88999*******999************************************************************* PP
GAGA_bind 246 vstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvti 300
+stkrrgaRiagrKmS+gafkk+LekLa+eGy+l+np+DLk++WAkHGtnkfvti
ONIVA10G01080.4 274 MSTKRRGARIAGRKMSHGAFKKVLEKLAGEGYNLNNPIDLKTFWAKHGTNKFVTI 328
******************************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF06217 | 3.4E-114 | 1 | 328 | IPR010409 | GAGA-binding transcriptional activator |
| SMART | SM01226 | 8.9E-192 | 1 | 329 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MDDDASMSIR WGGFFESPAR NLGLQLMSSV PADRDTKQLL SGSPFLHHQH QQQHVPHHHH 60 QPHHPRDCGA NGNANGGAMP PPPATEAPPS MPMNFQHHHP REHKVLHNLT VGHGSSHIAH 120 HDPVGYGMIP GTHTLQMMQQ QTEPQPQPPP PPQQPKEECI SSPLIEENVP VIDEPPPPKK 180 RQQGRQPKVP RAKKPKKSAA PREDGAPPNA PAPRRRGPRK NIGMVINGID LDLSRIPTPV 240 CSCTGAPQQC YRWGAGGWQS ACCTTTISTY PLPMSTKRRG ARIAGRKMSH GAFKKVLEKL 300 AGEGYNLNNP IDLKTFWAKH GTNKFVTISC LV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
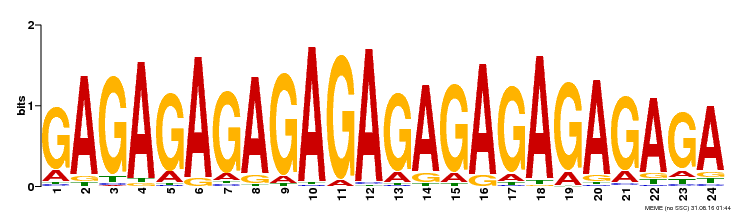
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012618 | 0.0 | CP012618.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 10 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015612725.1 | 0.0 | barley B recombinant-like protein A isoform X1 | ||||
| Refseq | XP_015612727.1 | 0.0 | barley B recombinant-like protein A isoform X1 | ||||
| Refseq | XP_015612730.1 | 0.0 | barley B recombinant-like protein B isoform X1 | ||||
| Refseq | XP_015612731.1 | 0.0 | barley B recombinant-like protein B isoform X1 | ||||
| Swissprot | P0DH88 | 0.0 | BBRA_ORYSJ; Barley B recombinant-like protein A | ||||
| Swissprot | P0DH89 | 0.0 | BBRB_ORYSJ; Barley B recombinant-like protein B | ||||
| TrEMBL | A0A0E0IP31 | 0.0 | A0A0E0IP31_ORYNI; Uncharacterized protein | ||||
| STRING | OGLUM10G00620.1 | 0.0 | (Oryza glumipatula) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14685.3 | 2e-59 | basic pentacysteine 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



