 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC01G08000.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 204aa MW: 23407.5 Da PI: 10.3865 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 38.8 | 2.2e-12 | 51 | 92 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +l +av+ + g++Wk+I Rt+ qc +rwqk+l
OPUNC01G08000.2 51 KGNWTPEEDAILSRAVQTYNGKNWKKI------DRTDVQCLHRWQKVL 92
79************************4......69**********986 PP
| |||||||
| 2 | Myb_DNA-binding | 63.7 | 3.6e-20 | 98 | 144 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd+++v+ v++lG++ W+tIa+ ++ gR +kqc++rw+++l
OPUNC01G08000.2 98 KGPWSKEEDDIIVQMVNKLGPKKWSTIAQALP-GRIGKQCRERWYNHL 144
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 52.1 | 1.5e-16 | 150 | 192 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE+ lv+a++++G++ W+ + ++ gRt++++k++w+
OPUNC01G08000.2 150 KEAWTQEEEITLVHAHRMYGNK-WAELTKFLP-GRTDNSIKNHWN 192
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.22 | 46 | 92 | IPR017930 | Myb domain |
| SMART | SM00717 | 6.8E-9 | 50 | 94 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.5E-10 | 51 | 92 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 7.59E-13 | 52 | 102 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-20 | 53 | 109 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 8.31E-10 | 53 | 92 | No hit | No description |
| PROSITE profile | PS51294 | 30.712 | 93 | 148 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.48E-31 | 95 | 191 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.7E-18 | 97 | 146 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.4E-18 | 98 | 144 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.80E-15 | 100 | 144 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-26 | 110 | 151 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.298 | 149 | 201 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.3E-12 | 149 | 197 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.9E-15 | 150 | 193 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-20 | 152 | 194 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.59E-10 | 152 | 192 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 204 aa Download sequence Send to blast |
MMTSDNGKAP DKSGESSGPP PAPQEGEIST EPKRRRPLNG RTTGPTRRST KGNWTPEEDA 60 ILSRAVQTYN GKNWKKIDRT DVQCLHRWQK VLNPELVKGP WSKEEDDIIV QMVNKLGPKK 120 WSTIAQALPG RIGKQCRERW YNHLNPGINK EAWTQEEEIT LVHAHRMYGN KWAELTKFLP 180 GRTDNSIKNH WNSSVYLPKF RVSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 5e-62 | 51 | 195 | 6 | 155 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 5e-62 | 51 | 195 | 6 | 155 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (PubMed:21862669). Transcription activator involved in the regulation of cytokinesis, probably via the activation of several G2/M phase-specific genes transcription (e.g. KNOLLE) (PubMed:17287251, PubMed:21862669). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). Required for the maintenance of diploidy (PubMed:21862669). {ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:21862669, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
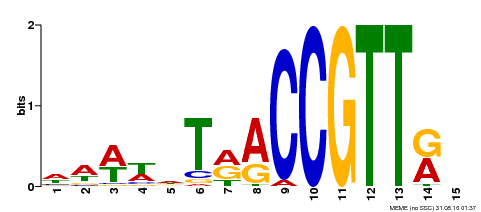
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Constant levels during cell cycle. Activated by CYCB1. {ECO:0000269|PubMed:17287251}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK318601 | 0.0 | AK318601.1 Oryza sativa Japonica Group cDNA, clone: J100066E24, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625869.1 | 1e-132 | transcription factor MYB3R-1 | ||||
| Swissprot | Q9S7G7 | 3e-89 | MB3R1_ARATH; Transcription factor MYB3R-1 | ||||
| TrEMBL | A0A0E0JFX9 | 1e-149 | A0A0E0JFX9_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC01G07980.1 | 1e-136 | (Oryza punctata) | ||||
| STRING | OPUNC01G08000.1 | 1e-136 | (Oryza punctata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32730.1 | 1e-91 | Homeodomain-like protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



