 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC01G08460.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 787aa MW: 84478.1 Da PI: 9.6027 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.9 | 1.6e-12 | 633 | 679 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
OPUNC01G08460.2 633 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYITDLQ 679
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 8.1E-50 | 229 | 414 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.831 | 629 | 678 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-18 | 632 | 696 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.34E-15 | 632 | 683 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 8.9E-18 | 633 | 692 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.1E-10 | 633 | 679 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.9E-16 | 635 | 684 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 787 aa Download sequence Send to blast |
MRYEQTTSPR AEKSPGVGDR AGSSTPRPPT SRVSSRVGSA RDPIPPGQGH PRSPFTPPLL 60 LLSLHITSRL PPPPPKSPRE PEKPEAPPPP PPPPPARAAN SGTLALAWGV EGRSGRRRRR 120 GCSDSATSGG GREGPASSFL GAPGRSGAPS VLDSVAAAPI GAREPQHGIM VMKMEVDEDG 180 ANGGTGGTWT DEDRALSASV LGTDAFAYLT KGGGAISEGL VAASLPVDLQ NRLQELVESD 240 RPGAGWNYAI FWQLSRTKSG DLVLGWGDGS CREPRDGEMG PAASAGSDEA KQRMRKRVLQ 300 RLHSAFGGVD EEDYAPGIDQ VTDTEMFFLA SMYFAFPRRA GGPGQVFAAG VPLWIPNTER 360 NVFPANYCYR GYLANAAGFR TIVLVPFESG VLELGSMQQV AESSDTLQTI RSVFAGAIGN 420 KAGVQRHEGN GPADKSPGLA KIFGKDLNLG RPSAGPGTGV SKADERSWEQ RTAGGGSSLL 480 PNVQRGLQNF TWSQARGLNS HQQKFGNGIL IVSNEATPRN NGVVDSPTAT QFQLQKASPL 540 QKLPQLQKPH QIVKPQQLVS QQQLQPQAPR QIDFSAGTSS KPGVLTKKPA GIDGEGAEVD 600 GLCKDEGPPP AIEDRRPRKR GRKPANGREE PLNHVEAERQ RREKLNQRFY ALRAVVPNIS 660 KMDKASLLGD AITYITDLQK KLKEMEVERE RLIESGMVDP RERTPRPEVD IQVVQDEVLV 720 RVMSPMENHP VRAIFQAFEE AEVHAGESKI TSNNGTAVHS FIIKCPGAEQ QTREKVIAAM 780 SRAMNSG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 2e-25 | 229 | 414 | 9 | 192 | Transcription factor MYC3 |
| 4rqw_B | 2e-25 | 229 | 414 | 9 | 192 | Transcription factor MYC3 |
| 4rs9_A | 2e-25 | 229 | 414 | 9 | 192 | Transcription factor MYC3 |
| 4yz6_A | 2e-25 | 229 | 414 | 9 | 192 | Transcription factor MYC3 |
| 5gnj_A | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 2e-26 | 627 | 689 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 614 | 622 | RRPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
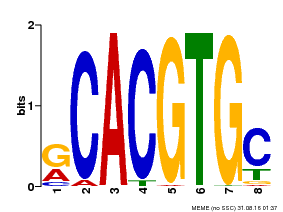
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CT833046 | 0.0 | CT833046.1 Oryza sativa (indica cultivar-group) cDNA clone:OSIGCRA221E23, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015621308.1 | 0.0 | transcription factor bHLH13 | ||||
| TrEMBL | A0A0E0JG40 | 0.0 | A0A0E0JG40_ORYPU; Uncharacterized protein | ||||
| STRING | ORUFI01G09400.1 | 0.0 | (Oryza rufipogon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 9e-70 | bHLH family protein | ||||



