 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC01G11630.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 555aa MW: 58625.6 Da PI: 6.3785 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.7 | 6.8e-16 | 306 | 352 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
OPUNC01G11630.2 306 VHNLSERRRRDRINEKMRALQELIPNC-----NKIDKASMLDEAIEYLKTLQ 352
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.09E-17 | 299 | 356 | No hit | No description |
| SuperFamily | SSF47459 | 1.57E-20 | 300 | 361 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.981 | 302 | 351 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.9E-20 | 306 | 360 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.3E-13 | 306 | 352 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 308 | 357 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 555 aa Download sequence Send to blast |
MTPAVHDFAS GFGATTQDNG DDDTVPWIHY PIIDDEDGAA PAALAAADYG SDFFSEPAAA 60 AAPPTDLGSL PASNHNGATN NKNAPVATTT TREPSKESHG GLSVPTTRAE PQPQLAADKL 120 PRSSRSGEGV MNFSLFSRPA VLARATLQSA QRPQGTDKAS NVTASNRVES TVVQTASGSR 180 SAPAFADQRA AAAWPPQPKE MPFTSTAAAP MPAAANLHHE MGRDMAGRAM PVHKTEARKA 240 PEATVATSSV CSGNGAGSDE LWRQQKRKCQ AQAECSASQD DDIDDEPGVL RRSGTRSTKR 300 SRTAEVHNLS ERRRRDRINE KMRALQELIP NCNKIDKASM LDEAIEYLKT LQLQVQMMSM 360 GTGLCIPPML LPTAMQHLQI PPMAHFPHLG MGLGYGMGVF DMSNTGALQM PPMPGAHFPC 420 PMIPGASPQG LGIPGTSTMP MFGVPGQTIP SSASSVPPFA SLAGLPVRPS GVPQVSSAMA 480 NMVQDQQQGI SNQQQQCLNK EAIQGANPGD PQMQIIMQGD NENFRIPSSA QTKSSQFSDG 540 TGKGTNARER DGAET |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 310 | 315 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
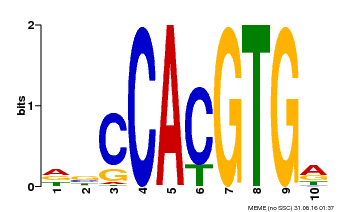
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK102252 | 0.0 | AK102252.1 Oryza sativa Japonica Group cDNA clone:J033088H11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015619034.1 | 0.0 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X2 | ||||
| Swissprot | Q0JNI9 | 0.0 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A0E0JH97 | 0.0 | A0A0E0JH97_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC01G11630.2 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 6e-35 | phytochrome interacting factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



