 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC02G02530.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 452aa MW: 49927.4 Da PI: 6.3984 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 104.3 | 7e-33 | 258 | 312 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWtpeLHerFv+av++L G+ekAtPk +l+lmkv+gLt++h+kSHLQkYRl
OPUNC02G02530.1 258 KPRLRWTPELHERFVDAVNKLEGPEKATPKGVLKLMKVEGLTIYHIKSHLQKYRL 312
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.818 | 255 | 315 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.79E-17 | 256 | 312 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-31 | 257 | 313 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.0E-25 | 258 | 313 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.9E-9 | 260 | 311 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 6.0E-22 | 346 | 392 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 452 aa Download sequence Send to blast |
MSTQSVIAVK QFSGSDKMAH TCTAPRPSAH ILSNANSDLC GSTNSSTSLP CSIQSSNIKT 60 ELISSSSLPK ILPFNAGSNP ENSHSRMSQH EFSDPILSSS SFFCTSLYTS SSMNSGSCRK 120 NSDLLFLPHP SKFEQQQNSA GQSSSSFLSN SGHVDDEHTD DVKDFLNLSS DGSFNGKSSA 180 MAYDEQMEFQ FLSEQLGIAI TNNEESPRLD DIYDRPPQLL SLPVSSCSDQ EDLQDAGSPA 240 KVQLSSSRSS SGTASCNKPR LRWTPELHER FVDAVNKLEG PEKATPKGVL KLMKVEGLTI 300 YHIKSHLQKY RLAKYLPETK EDKKQEEKKA KSVANGNDHV KKKSAQMAEA LRMQMEVQKQ 360 LHEQLEVQRQ LQLRIEEHAR YLQKILEEQQ KARESISSTM TSTTEGESPE FAPKEKTEDK 420 AETSSAPLSK YRISDTDAEC HSQVDNKKTA AG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-26 | 258 | 315 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phosphate starvation signaling (PubMed:26082401). Binds to P1BS, an imperfect palindromic sequence 5'-GNATATNC-3', to promote the expression of inorganic phosphate (Pi) starvation-responsive genes (PubMed:26082401). Functionally redundant with PHR1 and PHR2 in regulating Pi starvation response and Pi homeostasis (PubMed:26082401). {ECO:0000269|PubMed:26082401}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
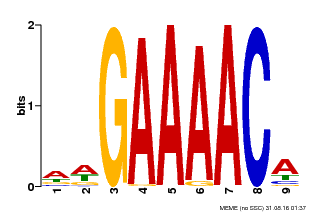
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated under Pi starvation conditions. {ECO:0000269|PubMed:26082401}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK065704 | 0.0 | AK065704.1 Oryza sativa Japonica Group cDNA clone:J013037C19, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015627390.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| Refseq | XP_015627391.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| Swissprot | Q6YXZ4 | 0.0 | PHR3_ORYSJ; Protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| TrEMBL | A0A0E0JVC5 | 0.0 | A0A0E0JVC5_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC02G02530.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4526 | 37 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 1e-61 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



