 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC02G13150.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 409aa MW: 44185.8 Da PI: 6.7998 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.7 | 9.6e-32 | 236 | 291 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rF++a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
OPUNC02G13150.1 236 KARRCWAPELHRRFLQALQQLGGSHVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 291
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.524 | 233 | 293 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.0E-28 | 234 | 294 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.73E-17 | 234 | 294 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.0E-27 | 236 | 291 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.8E-6 | 238 | 289 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 409 aa Download sequence Send to blast |
MEVDHADRDR ARRRCREYLF ALEEERRKIQ VFQRELPLCF DLVTQTIEGM RSQMDGAGSE 60 ETVSDQGPPP VLEEFIPLKP SLSLSSSEEE STHANAAKSG KKEEAETSER HSSPPPPPEA 120 KKVTPDWLQS VQLWSQEPPQ QPSSPSPTPA KDLPCKPVAL NAGKAGGAFQ PFEKEKRAEL 180 PASSTTAAAS STVVGDSGDK PTDDDTEKQM ETDKDNNKDA KDKDKDKEGQ SQPHRKARRC 240 WAPELHRRFL QALQQLGGSH VATPKQIREL MKVDGLTNDE VKSHLQKYRL HTRRPSTTGQ 300 SSATAGVAAP PAPQFVVVGS IWVPPPEYAA AAAAQQHVQL AAGNASGSAN PVYAPVAMLP 360 AGLQPHSHRK QQQQQGQRHS GSEGRQSGDA GDGSSSSPAV SSSSQTTSA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-13 | 236 | 289 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-13 | 236 | 289 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-13 | 236 | 289 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-13 | 236 | 289 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-13 | 236 | 289 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that may play a role in response to nitrogen. May be involved in a time-dependent signaling for transcriptional regulation of nitrate-responsive genes. Binds specifically to the DNA sequence motif 5'-GAATC-3' or 5'-GAATATTC-3'. Represses the activity of its own promoter trough binding to these motifs. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00160 | DAP | Transfer from AT1G25550 | Download |
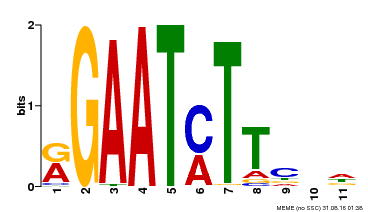
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by nitrate. {ECO:0000269|PubMed:23324170}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK064355 | 0.0 | AK064355.1 Oryza sativa Japonica Group cDNA clone:002-108-B01, full insert sequence. | |||
| GenBank | AK101809 | 0.0 | AK101809.1 Oryza sativa Japonica Group cDNA clone:J033066M05, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015623779.1 | 0.0 | transcription factor NIGT1 | ||||
| Swissprot | Q6Z869 | 0.0 | NIGT1_ORYSJ; Transcription factor NIGT1 | ||||
| TrEMBL | A0A0E0JZ84 | 0.0 | A0A0E0JZ84_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC02G13150.2 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6470 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 7e-63 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



