 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC02G35060.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 328aa MW: 34319.1 Da PI: 8.2321 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.2 | 0.00055 | 116 | 138 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y+C+ C+k+F++ L H +H
OPUNC02G35060.1 116 YECKTCNKCFPTFQALGGHRASH 138
99**********99999998887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 1.56E-8 | 115 | 138 | No hit | No description |
| Pfam | PF13912 | 1.9E-11 | 115 | 140 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.094 | 116 | 138 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.097 | 116 | 143 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 118 | 138 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.56E-8 | 212 | 239 | No hit | No description |
| Pfam | PF13912 | 9.1E-13 | 216 | 240 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.026 | 217 | 239 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.349 | 217 | 239 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 219 | 239 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MSVEDKHHLA AGVVKRKRTK RPRHHAAPAS SSESTTTEEE DMAHCLILLA QGAAVVDSKP 60 STPAPPPPPP PAQPPVLAAP APPPPAVVVK SERYTSRKYT EAATTADGVR AGFYVYECKT 120 CNKCFPTFQA LGGHRASHKK PRLAGADDDN ANATTNTNTN AIVVKSKPPL MTAPSPPSPP 180 PPQADAALCS VVVPDVTTVL SLNNVAASSM INKLRVHECS ICGAEFGSGQ ALGGHMRRHR 240 PLHAPPERAV TMATTTTAPD TKKAGSTSIN LELDLNLPAP SDEESVSPPP PPPPPPPPVL 300 LALDGQFDDG KKPILQLTTS AALVGCHY |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00487 | DAP | Transfer from AT5G04390 | Download |
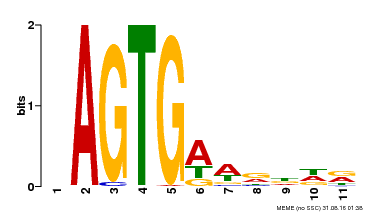
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP003989 | 0.0 | AP003989.4 Oryza sativa Japonica Group genomic DNA, chromosome 2, BAC clone:OJ1063_D06. | |||
| GenBank | AP004026 | 0.0 | AP004026.3 Oryza sativa Japonica Group genomic DNA, chromosome 2, BAC clone:OJ1136_C04. | |||
| GenBank | AP014958 | 0.0 | AP014958.1 Oryza sativa Japonica Group DNA, chromosome 2, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015623775.1 | 1e-140 | zinc finger protein ZAT5 | ||||
| TrEMBL | A0A0E0K739 | 0.0 | A0A0E0K739_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC02G35060.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2013 | 32 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G04390.1 | 5e-37 | C2H2 family protein | ||||



