 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC03G22220.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 485aa MW: 51412.1 Da PI: 4.9583 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.5 | 4.9e-16 | 55 | 102 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WTt Ed lv+ v+q+G g+W+++ r g+ R++k+c++rw ++l
OPUNC03G22220.1 55 KGPWTTAEDAVLVQHVRQHGEGNWNAVQRITGLLRCGKSCRLRWTNHL 102
79******************************99***********996 PP
| |||||||
| 2 | Myb_DNA-binding | 51 | 3.3e-16 | 108 | 151 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g+++++E++l++++++qlG++ W++ a++++ gRt++++k++w++
OPUNC03G22220.1 108 KGSFSPDEELLIAQLHAQLGNK-WARMASHLP-GRTDNEIKNYWNT 151
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.913 | 50 | 102 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.6E-13 | 54 | 104 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.06E-28 | 54 | 149 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.6E-13 | 55 | 102 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-20 | 56 | 109 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.97E-10 | 57 | 102 | No hit | No description |
| PROSITE profile | PS51294 | 22.55 | 103 | 157 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.0E-15 | 107 | 155 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.8E-15 | 108 | 151 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-24 | 110 | 156 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.83E-11 | 110 | 151 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 485 aa Download sequence Send to blast |
MEEMIKPAPA MSEEEKEVVA TAATGERRNE EATAEREEQE EEEEEAAAAP VVLKKGPWTT 60 AEDAVLVQHV RQHGEGNWNA VQRITGLLRC GKSCRLRWTN HLRPNLKKGS FSPDEELLIA 120 QLHAQLGNKW ARMASHLPGR TDNEIKNYWN TRTKRRQRAG LPVYPPDVQL HLAFAKRCRY 180 DDFSSPLSSP QQSAGSNVLT MDAADAAASA GYTSARPPPL DLAGQLAMGN RPVQFLSATP 240 FSAPSSPWGK PFARNAQFFQ FPHSSPVSPT TPTGPVHPVT PELSLGYGLH AGDRARLPPV 300 SPSPGARVEL PSSQLRPAMA PTASTTATAA GATGGLVGGA LQDHPNAASL EAMLQELHDA 360 IKIEPPALPE NRGTEGGGGG DMRGVSGDGK PEVELKDDIE TLFDLIIPAT FPAAPESAAA 420 TAASAAPNHS GSVSQHSSDD QDHSNADVVL DLPILTDGGG GSSEQDDWSL DGAACQWNNM 480 SGGIC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 2e-28 | 53 | 156 | 25 | 127 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
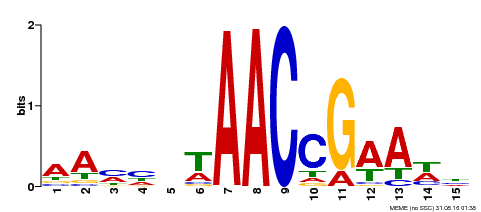
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC147962 | 0.0 | AC147962.1 Oryza sativa chromosome 3 BAC OSJNBa0083K01 genomic sequence, complete sequence. | |||
| GenBank | AP014959 | 0.0 | AP014959.1 Oryza sativa Japonica Group DNA, chromosome 3, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012611 | 0.0 | CP012611.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 3 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629365.1 | 0.0 | transcription factor MYB65 | ||||
| TrEMBL | A0A0E0KFS5 | 0.0 | A0A0E0KFS5_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC03G22220.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7420 | 37 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.1 | 5e-66 | myb domain protein 101 | ||||



