 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC03G29850.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 344aa MW: 37532 Da PI: 6.7382 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 27 | 7.4e-09 | 267 | 305 | 17 | 55 |
HHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 17 elFeknrypsaeereeLAkklgLterqVkvWFqNrRake 55
el +k +yps++++ LA+++gL+ +q+ +WF N+R ++
OPUNC03G29850.1 267 ELHYKWPYPSESQKVALAESTGLDLKQINNWFINQRKRH 305
5566789*****************************885 PP
| |||||||
| 2 | ELK | 37.2 | 6.3e-13 | 225 | 246 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKh+Ll+KYsgyL+sLkqE+s
OPUNC03G29850.1 225 ELKHHLLKKYSGYLSSLKQELS 246
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01256 | 4.3E-28 | 136 | 187 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 9.9E-23 | 142 | 185 | IPR005541 | KNOX2 |
| SMART | SM01188 | 1800 | 145 | 165 | IPR005539 | ELK domain |
| SMART | SM01188 | 4.9E-7 | 225 | 246 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.331 | 225 | 245 | IPR005539 | ELK domain |
| Pfam | PF03789 | 7.7E-10 | 225 | 246 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.617 | 245 | 308 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 3.21E-19 | 246 | 321 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 8.5E-13 | 247 | 312 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-27 | 250 | 309 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.50E-12 | 257 | 309 | No hit | No description |
| Pfam | PF05920 | 2.8E-16 | 265 | 304 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 283 | 306 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MEEISHHFGA VGASGGHGGH GHHQHQHHHP WGSSLSAIVA PPPPPQQTQA GGMAHMPLTL 60 NTAAAAVAAA GNGSGNPVLQ LANGSLLDAC GKAKEPSASA SASASYAADV GAPPEVAARL 120 TAVAQDLELR QRTALGGLGA ATEPELDQFM EAYHEMLVKY REELTRPLQE AMEFLRRVET 180 QLNTLSISGR SLRNILSSGS SEEDQEGSGG ETELPEIDAH GVDQELKHHL LKKYSGYLSS 240 LKQELSKKKK KGKLPKDARQ QLLNWWELHY KWPYPSESQK VALAESTGLD LKQINNWFIN 300 QRKRHWKPSD EMQFVMMDSY HPTNAAAFYM DGHFINDGGL YRLG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that regulates genes involved in development. May be involved in shoot formation during embryogenesis. Overexpression in transgenic plants causes altered leaf morphology (PubMed:10488233, PubMed:8755613, PubMed:9869405). Regulates anther dehiscence via direct repression of the auxin biosynthetic gene YUCCA4 (PubMed:29915329). Binds to the DNA sequence 5'-TGAC-3' in the promoter of the YUCCA4 gene and represses its activity during anther development (PubMed:29915329). Reduction of auxin levels at late stage of anther development, after meiosis of microspore mother cells, is necessary for normal anther dehiscence and seed setting (PubMed:29915329). {ECO:0000269|PubMed:10488233, ECO:0000269|PubMed:29915329, ECO:0000269|PubMed:8755613, ECO:0000269|PubMed:9869405}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
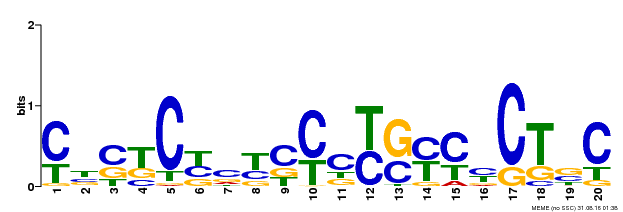
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FJ940206 | 0.0 | FJ940206.1 Oryza sativa Japonica Group clone KCB522D03 homeobox protein mRNA, complete cds. | |||
| GenBank | RICOSH1 | 0.0 | D16507.1 Oryza sativa Japonica Group OSH1 mRNA for homeobox protein, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629392.1 | 0.0 | homeobox protein knotted-1-like 6 | ||||
| Swissprot | P46609 | 0.0 | KNOS6_ORYSJ; Homeobox protein knotted-1-like 6 | ||||
| TrEMBL | A0A0E0KII0 | 0.0 | A0A0E0KII0_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC03G29850.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9886 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 8e-79 | KNOTTED-like from Arabidopsis thaliana | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



