 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC05G01460.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 303aa MW: 34693.6 Da PI: 8.642 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 20.1 | 1.7e-06 | 31 | 55 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ C+ s+srk++L+rH+ tH
OPUNC05G01460.1 31 FSCHvdGCPFSYSRKDHLNRHLLTH 55
89*********************99 PP
| |||||||
| 2 | zf-C2H2 | 13.4 | 0.00022 | 60 | 85 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+ Cp C++ F+ k n++rH + H
OPUNC05G01460.1 60 FACPveGCNRKFTIKGNMQRHVQEmH 85
78*******************99877 PP
| |||||||
| 3 | zf-C2H2 | 21.2 | 7.8e-07 | 97 | 121 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp +Cgk+F+ s L++H +H
OPUNC05G01460.1 97 FICPeeNCGKTFKYASKLQKHEESH 121
78*******************9988 PP
| |||||||
| 4 | zf-C2H2 | 17.2 | 1.4e-05 | 188 | 212 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
+C+ dC+ sFs ksnL +H + H
OPUNC05G01460.1 188 RCHfkDCKLSFSKKSNLDKHVKAvH 212
7*******************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 2.0E-9 | 28 | 57 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.26E-6 | 28 | 57 | No hit | No description |
| SMART | SM00355 | 0.0016 | 31 | 55 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.349 | 31 | 60 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 33 | 55 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.12E-6 | 58 | 86 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 7.1E-8 | 58 | 86 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 3.8E-4 | 60 | 85 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.887 | 60 | 90 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 62 | 85 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-7 | 93 | 121 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.57E-5 | 94 | 123 | No hit | No description |
| SMART | SM00355 | 0.0018 | 97 | 121 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.24 | 97 | 121 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 99 | 121 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 12 | 129 | 154 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 131 | 154 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 13 | 157 | 178 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 1.72E-7 | 168 | 229 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.8E-5 | 168 | 209 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.016 | 187 | 212 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.071 | 187 | 217 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 189 | 212 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.4E-6 | 211 | 240 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 8.974 | 218 | 249 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 3.5 | 218 | 244 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 220 | 244 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 303 aa Download sequence Send to blast |
MALNGLVSGW GDSPEKLHQE CLVHTLFQRP FSCHVDGCPF SYSRKDHLNR HLLTHQGKLF 60 ACPVEGCNRK FTIKGNMQRH VQEMHKDGSP CESKKEFICP EENCGKTFKY ASKLQKHEES 120 HVKLDYSEVI CCEPGCMKAF TNLECLKAHN ESCHRHVVCD VCGTKQLKKN FKRHQRMHEG 180 SCVTERIRCH FKDCKLSFSK KSNLDKHVKA VHEQKRPFVC GFSGCGKSFS YKHVRDNHEK 240 SSAHVYVQAN FEEIDGERPR QAGGRKRKAI PVDSLMRKRV AAPDDDSPAC DDGTEYLRWL 300 LSG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 7e-14 | 28 | 149 | 40 | 155 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 7e-14 | 28 | 149 | 40 | 155 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
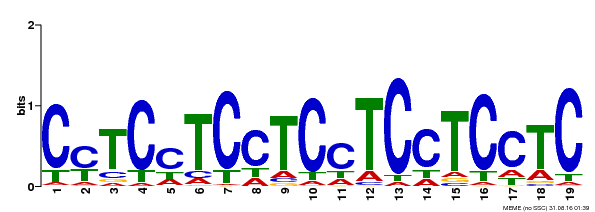
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK101631 | 0.0 | AK101631.1 Oryza sativa Japonica Group cDNA clone:J033054O17, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015637660.1 | 0.0 | transcription factor IIIA isoform X1 | ||||
| Refseq | XP_015637661.1 | 0.0 | transcription factor IIIA isoform X2 | ||||
| Swissprot | Q84MZ4 | 3e-88 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0E0KXX0 | 0.0 | A0A0E0KXX0_ORYPU; Uncharacterized protein | ||||
| STRING | OBART05G01510.1 | 0.0 | (Oryza barthii) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 2e-90 | transcription factor IIIA | ||||



