 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC05G01460.4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 218aa MW: 23714.9 Da PI: 7.2365 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 22.1 | 3.9e-07 | 156 | 176 | 3 | 23 |
ETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 CpdCgksFsrksnLkrHirtH 23
C+ Cg sF+ + +Lk+H+++H
OPUNC05G01460.4 156 CKVCGASFKKPAHLKQHMQSH 176
*******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 15 | 107 | 129 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.01 | 154 | 176 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.05 | 154 | 181 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.9E-6 | 156 | 185 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 4.36E-6 | 156 | 185 | No hit | No description |
| PROSITE pattern | PS00028 | 0 | 156 | 176 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF12874 | 9.5E-5 | 156 | 176 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 218 aa Download sequence Send to blast |
MANGGGTVEG EDEGTRAGLR WYPNPAPAFS NPNPLRLAYS ATATTAWFES RCAAVANIWS 60 LAEPLIQRIP LQLEMGSVEL GTEEGEVAGG EGESKGAAPP ARDIRRYKCD FCSVVRSKKG 120 LIRAHVLEHH KDEVDDLDDY FGRGGGKSHK GIDRDCKVCG ASFKKPAHLK QHMQSHSLER 180 TPKCEKALSG LVPGWGDPSG KLLQECLFHT LLQTIVHI |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
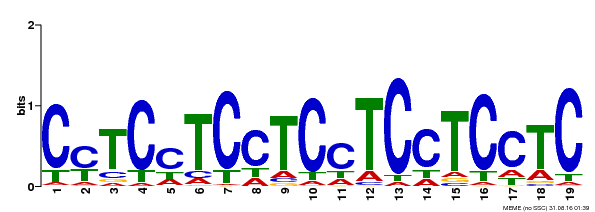
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK099528 | 1e-179 | AK099528.1 Oryza sativa Japonica Group cDNA clone:J013031H23, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015637660.1 | 2e-59 | transcription factor IIIA isoform X1 | ||||
| TrEMBL | A0A0E0KXX3 | 1e-161 | A0A0E0KXX3_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC05G01460.3 | 1e-128 | (Oryza punctata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 4e-17 | transcription factor IIIA | ||||



