 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC05G16800.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 619aa MW: 68232.8 Da PI: 8.6423 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 26.8 | 1.2e-08 | 64 | 101 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g WT+eEde+l +av + g++Wk+I qc +rwqk+
OPUNC05G16800.1 64 KGGWTPEEDEKLRKAVDIYNGKNWKKI----E-----VQCLHRWQKV 101
688***********************5....3.....4577777775 PP
| |||||||
| 2 | Myb_DNA-binding | 58.2 | 1.9e-18 | 108 | 154 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT++Ed+ ++++vk++G++ W+ Iar ++ gR +kqc++rw+++l
OPUNC05G16800.1 108 KGPWTQQEDDVIINLVKKHGPKKWSVIARSLN-GRIGKQCRERWHNHL 154
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 51.5 | 2.3e-16 | 160 | 203 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+ +WT eE+ l++a+ ++G++ W+ Ia+ ++ gRt++++k++w++
OPUNC05G16800.1 160 KEAWTVEEEHVLARAHCMYGNK-WAEIAKLLP-GRTDNSIKNHWNS 203
579*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 7.082 | 59 | 102 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.08E-11 | 62 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-4 | 63 | 104 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.5E-6 | 64 | 101 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-13 | 66 | 109 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 30.357 | 103 | 158 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.24E-31 | 105 | 201 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-16 | 107 | 156 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.3E-17 | 108 | 154 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.75E-15 | 110 | 154 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 9.6E-25 | 110 | 157 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.6E-22 | 158 | 209 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.3E-13 | 159 | 207 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 21.01 | 159 | 209 | IPR017930 | Myb domain |
| Pfam | PF00249 | 5.3E-15 | 160 | 203 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.14E-10 | 162 | 205 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 619 aa Download sequence Send to blast |
MPAVKVEEEE EEEEEVEERN PVASSPSVSE GSAHAALASP VAATTPADSV FGRRRKSGPV 60 RRAKGGWTPE EDEKLRKAVD IYNGKNWKKI EVQCLHRWQK VLDPELIKGP WTQQEDDVII 120 NLVKKHGPKK WSVIARSLNG RIGKQCRERW HNHLDPQIRK EAWTVEEEHV LARAHCMYGN 180 KWAEIAKLLP GRTDNSIKNH WNSSLRKKMD DYNTRNILPL QPPVVGDDLK QLPKRPPADN 240 HFDLNKEPII CSRDRLGVVH SDPTSHQRAS NLKYFKGCAD YLSLGQPVTS CEASAADDSA 300 FDLATQGMRM DSVHDKGTGN NFVCGKVQGI NFLGDKGLKI NQISDKMGCS RQAKREGEAA 360 INSGGSSLQS EAHSVGSLCY QIPKMEDIAP AESPVFTANY VPEHSRNVMH SPNGYTTPSP 420 THGKGSDQLS VESILRSAAE KFHGTPSILR RRKRDKPTPA EDNDLKIGRL SSDDFHTPIG 480 KSTTDSPQSF KTAALLSLGP MDGQGFSTAS GSLDVSPPYR LRSKRLAVLK TVQNHLDFSS 540 DEMSICDTTM KSACGNSDCA NASSGVSSIQ GKKLDEHMIG LETLTMNFAH TTKTTTNVLK 600 WARRPDASKL NVVTVPKGK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 8e-63 | 63 | 209 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 8e-63 | 63 | 209 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 449 | 456 | RRRKRDKP |
| 2 | 450 | 455 | RRKRDK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in abiotic stress responses (PubMed:17293435, PubMed:19279197). May play a regulatory role in tolerance to salt, cold, and drought stresses (PubMed:17293435). Transcriptional activator that binds specifically to a mitosis-specific activator cis-element 5'-(T/C)C(T/C)AACGG(T/C)(T/C)A-3', found in promoters of cyclin genes such as CYCB1-1 and KNOLLE (AC Q84R43). Positively regulates a subset of G2/M phase-specific genes, including CYCB1-1, CYCB2-1, CYCB2-2, and CDC20.1 in response to cold treatment (PubMed:19279197). {ECO:0000269|PubMed:17293435, ECO:0000269|PubMed:19279197}. | |||||
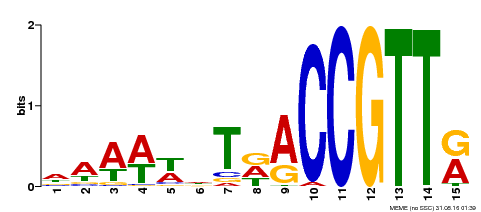
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by cold, drought and salt stresses. {ECO:0000269|PubMed:17293435}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK106756 | 0.0 | AK106756.1 Oryza sativa Japonica Group cDNA clone:002-115-D07, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015637435.1 | 0.0 | transcription factor MYB3R-2-like | ||||
| Swissprot | Q0JHU7 | 0.0 | MB3R2_ORYSJ; Transcription factor MYB3R-2 | ||||
| TrEMBL | A0A0E0L3E2 | 0.0 | A0A0E0L3E2_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC05G16800.2 | 0.0 | (Oryza punctata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 4e-95 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



