 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC05G18630.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 411aa MW: 46035.3 Da PI: 7.1082 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 107.2 | 9.1e-34 | 44 | 98 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+WtpeLH+rFv+av+qLGG+ekAtPkt+++lm+++gLtl+h+kSHLQkYRl
OPUNC05G18630.1 44 KPRLKWTPELHQRFVDAVNQLGGAEKATPKTVMRLMGIPGLTLYHLKSHLQKYRL 98
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.387 | 41 | 101 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-32 | 42 | 99 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 8.6E-17 | 43 | 99 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.2E-23 | 44 | 99 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.7E-10 | 46 | 97 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 2.8E-22 | 145 | 192 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 411 aa Download sequence Send to blast |
MYHQHQGRSD LFTTRTSFPM ERHLFLHGGN TQGDSGLVLS TDAKPRLKWT PELHQRFVDA 60 VNQLGGAEKA TPKTVMRLMG IPGLTLYHLK SHLQKYRLSK NLQGQANVGT TKNALGCTAV 120 VDRIPGTSAL AMASANAIPQ AEKTIQIGEA LQMQIEVQRQ LNEQLEVQRH LQLRIEAQGK 180 YLQAVLEQAQ ETLGKQNLGP ASLEDAKIKI SELVSQVSNE CLSNAVTEIR ESSSIHRLEP 240 RQIQFVESSA NNSLTAAEGF KEHRLQNHGV LKAYDDSTLF CRKQSQDQES QYSLNRSLSE 300 RRMGHLYNGK QYHKAEGSDS DTEVLHEYIT PQKNGGGSTT SSTSGSKEIN VEKLYLDEPS 360 CKRQAVDYQR ESKLLDFDQQ SSGKNLDLNT HNIDDNDQGY RHFDLNGFSW S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-20 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-20 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-20 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-20 | 44 | 100 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-20 | 43 | 100 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
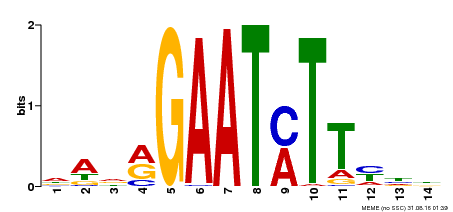
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK241362 | 0.0 | AK241362.1 Oryza sativa Japonica Group cDNA, clone: J065151H17, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015640413.1 | 0.0 | myb-related protein 2 isoform X1 | ||||
| Refseq | XP_015640414.1 | 0.0 | myb-related protein 2 isoform X1 | ||||
| Refseq | XP_015640415.1 | 0.0 | myb-related protein 2 isoform X1 | ||||
| TrEMBL | A0A0E0L422 | 0.0 | A0A0E0L422_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC05G18630.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12279 | 31 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 7e-98 | G2-like family protein | ||||



