 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC07G13970.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 397aa MW: 41420.7 Da PI: 6.7865 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.2 | 3.9e-18 | 13 | 60 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg W++eEde+lv+++ ++G +W++++++ g+ R++k+c++rw +yl
OPUNC07G13970.1 13 RGLWSPEEDEKLVRYISEHGHSCWSSVPKHAGLQRCGKSCRLRWINYL 60
788*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 50.5 | 4.8e-16 | 66 | 109 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg ++++E+ ++d+++ lG++ W+ Ia++++ gRt++++k++w++
OPUNC07G13970.1 66 RGTFSEQEERTIIDVHRILGNR-WAQIAKHLP-GRTDNEVKNFWNS 109
899*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.6E-27 | 4 | 63 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.005 | 8 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.24E-28 | 10 | 107 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-14 | 12 | 62 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-16 | 13 | 60 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.59E-13 | 16 | 60 | No hit | No description |
| PROSITE profile | PS51294 | 25.597 | 61 | 115 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-24 | 64 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.4E-15 | 65 | 113 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.4E-15 | 66 | 109 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.33E-11 | 68 | 108 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 397 aa Download sequence Send to blast |
MGHHCCSKQK VKRGLWSPEE DEKLVRYISE HGHSCWSSVP KHAGLQRCGK SCRLRWINYL 60 RPDLKRGTFS EQEERTIIDV HRILGNRWAQ IAKHLPGRTD NEVKNFWNSC IKKKLIAQGL 120 DPKTHNLLPA SKTLLHGVGG AANPSGNGLA QFQSNNGVAA AGTTPFTISS PAKAAAFDVA 180 PPAIPPALYD VVLPANPAGG MLMAHDHHHH QAAAAAAVVA YPYADHGNGG GVLMSFRDQN 240 ANVHGAASMD FMNGSSSSSS MEQHGGGGMS NGNGFSASMA AFMDEEAAMW ATAVEPPGMG 300 GLAGMDQVAQ QQQVLVQDAA VGVGPTTLMM HGGAATGAGA MVDKSVEMVD VSSAVYGGAG 360 GATATAFDLD LMVESCGMFC GGGGGAGNAM EQLQWDC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-34 | 13 | 115 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00350 | DAP | Transfer from AT3G12720 | Download |
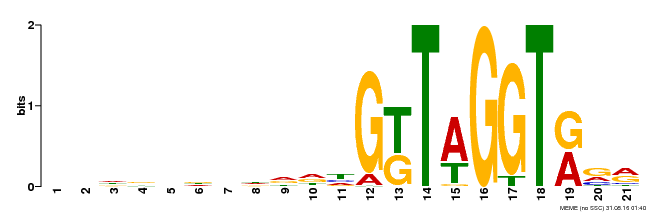
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012615 | 0.0 | CP012615.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 7 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015645809.1 | 0.0 | myb-related protein Hv33 | ||||
| TrEMBL | A0A0E0LKX6 | 0.0 | A0A0E0LKX6_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC07G13970.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9751 | 35 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12720.1 | 1e-82 | myb domain protein 67 | ||||



