 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OPUNC10G08850.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 449aa MW: 47802.4 Da PI: 6.7279 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.7 | 2.1e-05 | 102 | 124 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ C+k F r nL+ H r H
OPUNC10G08850.1 102 FVCEVCNKGFQRDQNLQLHRRGH 124
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 13.1 | 0.00029 | 170 | 192 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cgk++ +s+ k H +++
OPUNC10G08850.1 170 WRCERCGKRYAVHSDWKAHVKNC 192
58******************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 9.3E-6 | 101 | 124 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 5.09E-8 | 101 | 124 | No hit | No description |
| SMART | SM00355 | 0.021 | 102 | 124 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF12171 | 8.4E-5 | 102 | 124 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE profile | PS50157 | 10.97 | 102 | 124 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 104 | 124 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.6E-4 | 158 | 190 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 5.09E-8 | 165 | 190 | No hit | No description |
| SMART | SM00355 | 99 | 170 | 190 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 449 aa Download sequence Send to blast |
MLLSDLSSDQ EATGSNSHGG GGGGDHMVVG SHGAHVVLSN LFLPPVATTM LLPPAVMVRP 60 AAMAVVEEEP KAKKKRSLPG NPDPEAEVIA LSPRALVATN RFVCEVCNKG FQRDQNLQLH 120 RRGHNLPCGD GGAGAEEEGP TCVHHDPARA LGDLTGIKKH FSRKHGEKRW RCERCGKRYA 180 VHSDWKAHVK NCGTREYRCD CGILFSRKDS LLTHRAFCDA LAEESARLLA AANNSGSITT 240 CNNNSSSNIN SISNNNNLLI TSSSSSSPPL FLPFSTTPPA AENPNPNPLL FLQQHQAAHH 300 HQLLPQFQLP SSPPAYFDHL AFGAGGVITG SSNDDNSSIA GDVMVAGDSV SFGLTSEGSV 360 TMHAGVGRRL TRDFLGVDHD AGEVEELELD HELPLSTTTA TATACQGCSF AATTTAVSRA 420 ACCATDFTTG SRQYLGRLPP VNETWSHNF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 6e-33 | 166 | 235 | 3 | 72 | Zinc finger protein JACKDAW |
| 5b3h_F | 6e-33 | 166 | 235 | 3 | 72 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that acts as a flowering master switch in both long and short days, independently of the circadian clock (PubMed:18790997, PubMed:18725639, PubMed:18774969, PubMed:19304997, PubMed:24280027). Promotes flowering upstream of HD1 by up-regulating FTL1, FTL4, FTL5, FTL6, EHD1, HD3A and RFT1 (PubMed:18790997, PubMed:18725639, PubMed:18774969). Seems to repress FTL11 expression (PubMed:18725639). May recognize the consensus motif 5'-TTTGTCGTAAT-3' in target gene promoters (PubMed:18725639). {ECO:0000269|PubMed:18725639, ECO:0000269|PubMed:18774969, ECO:0000269|PubMed:18790997, ECO:0000269|PubMed:24280027, ECO:0000303|PubMed:19304997}. | |||||
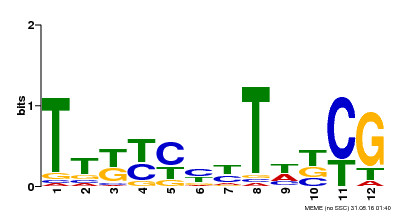
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00673 | SELEX | Transfer from GRMZM2G011357 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by FKF1 (PubMed:25850808). Down-regulated by DLF1 (PubMed:25036785). During the basic vegetative growth phase (BVP, photoperiod-insensitive phase), suppressed via phytochrome-mediated light signals involving the phytochromobilin synthase HY2 (PubMed:25573482). {ECO:0000269|PubMed:25036785, ECO:0000269|PubMed:25573482, ECO:0000269|PubMed:25850808}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB359195 | 1e-153 | AB359195.1 Oryza sativa Japonica Group Ehd2 gene for early heading date 2, complete cds, cultivar: Tohoku IL9. | |||
| GenBank | AB359196 | 1e-153 | AB359196.1 Oryza sativa Japonica Group Ehd2 mRNA for early heading date 2, complete cds, cultivar: Tohoku IL9. | |||
| GenBank | AB359197 | 1e-153 | AB359197.1 Oryza sativa Japonica Group Ehd2 gene for early heading date 2, complete cds, cultivar: R0364. | |||
| GenBank | AB359198 | 1e-153 | AB359198.1 Oryza sativa Japonica Group Ehd2 mRNA for early heading date 2, complete cds, cultivar: R0364. | |||
| GenBank | AC027658 | 1e-153 | AC027658.1 Oryza sativa (japonica cultivar-group) BAC nbxb0006I13 chromosome 10, complete sequence. | |||
| GenBank | AP014966 | 1e-153 | AP014966.1 Oryza sativa Japonica Group DNA, chromosome 10, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012618 | 1e-153 | CP012618.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 10 sequence. | |||
| GenBank | FJ009578 | 1e-153 | FJ009578.1 Oryza sativa Japonica Group cultivar Zhonghua 11 Cys2/His2-type zinc finger transcription factor (RID1) mRNA, complete cds. | |||
| GenBank | FJ009579 | 1e-153 | FJ009579.1 Oryza sativa Japonica Group cultivar Zhonghua 11 Cys2/His2-type zinc finger transcription factor (RID1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015614407.1 | 0.0 | protein indeterminate-domain 12 | ||||
| Swissprot | B1B534 | 0.0 | EHD2_ORYSJ; Protein EARLY HEADING DATE 2 | ||||
| TrEMBL | A0A0E0M7R6 | 0.0 | A0A0E0M7R6_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC10G08850.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP13096 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G45260.1 | 1e-76 | C2H2-like zinc finger protein | ||||



