 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ORGLA03G0326600.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 658aa MW: 68282.6 Da PI: 7.2929 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 39 | 2e-12 | 309 | 365 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng.krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g rk + g ++ +e+Aa+a++ a++k++g
ORGLA03G0326600.1 309 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqTRKGRQ-GGYDMEEKAARAYDLAALKYWG 365
78*******************77776664346655.77******************98 PP
| |||||||
| 2 | AP2 | 47.9 | 3.4e-15 | 408 | 459 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I +s +k +lg+f t eeAa+a++ a+ k++g
ORGLA03G0326600.1 408 SIYRGVTRHHQHGRWQARIGRVSG---NKDLYLGTFSTQEEAAEAYDVAAIKFRG 459
57****************999754...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 2.55E-14 | 309 | 374 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.2E-9 | 309 | 365 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.03E-16 | 309 | 373 | No hit | No description |
| PROSITE profile | PS51032 | 17.359 | 310 | 373 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.4E-12 | 310 | 374 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.3E-23 | 310 | 379 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-6 | 311 | 322 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-18 | 408 | 468 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 1.7E-10 | 408 | 459 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.67E-23 | 408 | 469 | No hit | No description |
| PROSITE profile | PS51032 | 19.137 | 409 | 467 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-18 | 409 | 467 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.9E-34 | 409 | 473 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-6 | 449 | 469 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 658 aa Download sequence Send to blast |
MASGNSSSSS GSMAATAGGV GGWLGFSLSP HMATHCAGGV DDVGHHHHHH VHQHQQQHGG 60 GLFYNPAAVA SSFYYGGGHD AVVTSAAGGG SYYGAGFSSM PLKSDGSLCI MEALRGGDQE 120 QQGVVVSASP KLEDFLGAGP AMALSLDNSA FYYGGHGHHQ GHAQDGGAVG GDPHHGGGGF 180 LQCAVIPGAG AGHDAAYAHA ALVHDQSAAA MAAGWAAMHG GGYDIANAAA DDVCAAGPII 240 PTGGHLHPLT LSMSSAGSQS SCVTVQAAAA GEPYMAMDAV SKKRGGADRA GQKQPVHRKS 300 IDTFGQRTSQ YRGVTRHRWT GRYEAHLWDN SCKKEGQTRK GRQGGYDMEE KAARAYDLAA 360 LKYWGPSTHI NFPLEDYQEE LEEMKNMSRQ EYVAHLRRKS SGFSRGASIY RGVTRHHQHG 420 RWQARIGRVS GNKDLYLGTF STQEEAAEAY DVAAIKFRGL NAVTNFDITR YDVDKILESS 480 TLLPGELARR KGKVGDGGGA AAVADAAAAL VQAGNVAEWK MATAAALPAA ARTEQQQQHG 540 HGGHQHHDLL PSDAFSVLQD IVSTVDAAGA PPRAPHMSMA ATSLGNSREQ SPDRGVGVGG 600 GGGGVLATLF AKPAAASKLY SPVPLNTWAS PSPAVSSVPA RAGVSIAHLP MFTAWTDA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as positive regulator of adventitious (crown) root formation by promoting its initiation. Promotes adventitious root initiation through repression of cytokinin signaling by positively regulating the two-component response regulator RR1. Regulated by the auxin response factor and transcriptional activator ARF23/ARF1. {ECO:0000269|PubMed:21481033}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
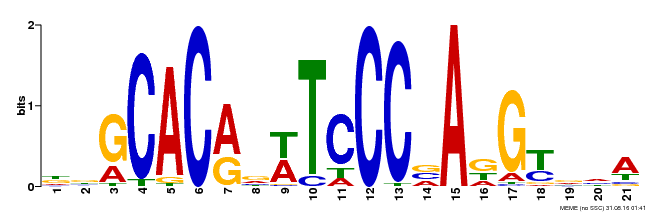
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | ORGLA03G0326600.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by auxin. {ECO:0000269|PubMed:21481033}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK241712 | 0.0 | AK241712.1 Oryza sativa Japonica Group cDNA, clone: J065197H24, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025879550.1 | 0.0 | AP2-like ethylene-responsive transcription factor CRL5 isoform X2 | ||||
| Swissprot | Q84Z02 | 1e-159 | CRL5_ORYSJ; AP2-like ethylene-responsive transcription factor CRL5 | ||||
| TrEMBL | I1PFX0 | 0.0 | I1PFX0_ORYGL; Uncharacterized protein | ||||
| STRING | ORGLA03G0326600.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP948 | 38 | 133 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-129 | AP2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



