 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ORUFI01G28580.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 563aa MW: 59311.8 Da PI: 9.3903 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 104.8 | 4.5e-33 | 353 | 410 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt+++Cpvkk+vers +dp vv++tYeg+H+h+
ORUFI01G28580.1 353 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCPVKKRVERSYQDPAVVITTYEGKHTHP 410
8********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.3E-35 | 337 | 412 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.96E-29 | 344 | 412 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.535 | 347 | 412 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.6E-39 | 352 | 411 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-26 | 353 | 410 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 563 aa Download sequence Send to blast |
MGVAVHWRRA GDSLHMGGEP RARAEAAWAA ALPAALVALV RDTATYTRMM HRLRMETPRG 60 PTCQLDPCCC CCTAHVIPPP PPVSRTHATR RDAEASVPPP PASAAVSSRS DGTGQMAAGV 120 TLACAAPPPL RAPRASDGGR RRGVVKGGAG TDTCRSPQRL NVRPRERERV RACVRARAKN 180 HEHGQRRREA AVDPAMSGEY QFQDELAPLF ARPGGGAGEM QMLPSSWFAD YLQAGTPMQM 240 DYDLMCRALE LPVGEDVKRE VGVVDVVAAG GGGGGERGGG GGGAGAGAGE EESPARCKKE 300 EDENKEEGKG EEDEGHKNKK GSAAKGGKAG KGEKRARQPR FAFMTKSEVD HLEDGYRWRK 360 YGQKAVKNSP YPRSYYRCTT QKCPVKKRVE RSYQDPAVVI TTYEGKHTHP IPATLRGSTH 420 LLAAHAQAAA AAAAAHQLHH HHGHHGHHGM APPLPLGSGA TAQFGRSSGI DVLSSFLPRA 480 AAAHHGITTM GGAAATTTTS HGLNSAISGG GGVSSETTSA VTVAASAQPS SPAALQMQHF 540 MAQDLGLLQD MLLPSFIHGT NQP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 4e-27 | 343 | 413 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 4e-27 | 343 | 413 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
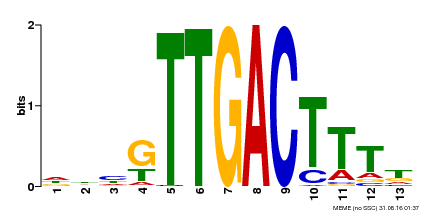
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY341844 | 0.0 | AY341844.1 Oryza sativa (japonica cultivar-group) WRKY3 mRNA, complete cds. | |||
| GenBank | AY341855 | 0.0 | AY341855.1 Oryza sativa (japonica cultivar-group) WRKY14 mRNA, complete cds. | |||
| GenBank | AY870604 | 0.0 | AY870604.1 Oryza sativa (japonica cultivar-group) WRKY16 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015640235.1 | 0.0 | uncharacterized protein LOC4326856 isoform X1 | ||||
| TrEMBL | A0A0E0N0E1 | 0.0 | A0A0E0N0E1_ORYRU; Uncharacterized protein | ||||
| STRING | ORUFI01G28580.1 | 0.0 | (Oryza rufipogon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP623 | 37 | 173 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G29860.1 | 1e-47 | WRKY DNA-binding protein 71 | ||||



