 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ORUFI05G17320.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 838aa MW: 91790.2 Da PI: 6.1924 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.4 | 5.6e-35 | 167 | 241 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
ORUFI05G17320.1 167 CQVPGCEADIRELKGYHRRHRVCLRCAHAAAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 241
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.5E-27 | 161 | 228 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.487 | 164 | 241 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.35E-33 | 165 | 244 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.8E-27 | 167 | 241 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 838 aa Download sequence Send to blast |
MDAPGGGGGG GGVDAGEPVW DWGNLLDFAV HDDDSLVLPW GDDSIGIEAD PAEAALLPPA 60 PSPQPAEAEA EAAGPASLPS SMQAEGSKRR VRKRDPRLVC PNYLAGRVPC ACPEIDEMAA 120 ALEVEDVATE LLAGARKKPK GAGRGSGAAV GGSGGGASRG TPAEMKCQVP GCEADIRELK 180 GYHRRHRVCL RCAHAAAVML DGVQKRYCQQ CGKFHILLDF DEDKRSCRRK LERHNKRRRR 240 KPDSKGILEK DIDDQLDFSA DGSGDGELRE ENIDVTTSET LETVLSNKVL DRETPVGSDD 300 VLSSPTCAQP SLQIDQSKSL VTFAASVEAC LGTKQENTKL TNSPVHDTKS TYSSSCPTGR 360 VSFKLYDWNP AEFPRRLRHQ IFEWLSSMPV ELEGYIRPGC TILTVFVAMP QHMWDKLSED 420 TGNLVKSLVN APNSLLLGKG AFFIHVNNMI FQVLKDGATL TSTRLEVQSP RIHYVHPSWF 480 EAGKPIDLIL CGSSLDQPKF RSLVSFDGLY LKHDCRRILS HETFDCIGSG EHILDSQHEI 540 FRINITTSKL DTHGPAFVEV ENMFGLSNFV PILVGSKHLC SELEQIHDAL CGSSDISSDP 600 CELRGLRQTA MLGFLIDIGW LIRKPSIDEF QNLLSLANIQ RWICMMKFLI QNDFINVLEI 660 IVNSLDNIIG SELLSNLEKG RLENHVTEFL GYVSEARNIV DNRPKYDKQR QVDTRWAGDY 720 APNQPKLGIS VPLAESTGTS GEHDLHSTNA ASGEEENMPL VTKALPHRQC CHPETSARWL 780 NAASIGAFPG GAMRMRLATT VVIGAVVCFA ACVVLFHPHR VGVLAAPVKR YLSRNYSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 3e-37 | 166 | 246 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 223 | 241 | KRSCRRKLERHNKRRRRKP |
| 2 | 227 | 239 | RRKLERHNKRRRR |
| 3 | 234 | 241 | NKRRRRKP |
| 4 | 235 | 240 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
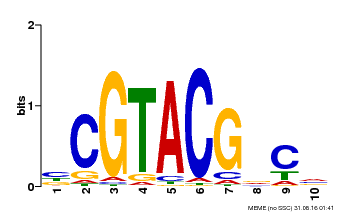
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK100057 | 0.0 | AK100057.1 Oryza sativa Japonica Group cDNA clone:J013158B16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015640052.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A0E0PME1 | 0.0 | A0A0E0PME1_ORYRU; Uncharacterized protein | ||||
| STRING | ORUFI05G17320.1 | 0.0 | (Oryza rufipogon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-144 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



