 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | ORUFI07G23760.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1032aa MW: 115058 Da PI: 5.7325 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 180.4 | 2.1e-56 | 21 | 137 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+ ++rwl+++ei++iL n++k++++ e+++rp sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+v+vl+cyYah+een++fq
ORUFI07G23760.1 21 LEAQNRWLRPTEICHILSNYKKFSIAPEPPNRPASGSLFLFDRKILRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSVDVLHCYYAHGEENENFQ 114
5679****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rr+ywlLee + +ivlvhylevk
ORUFI07G23760.1 115 RRTYWLLEEGFMNIVLVHYLEVK 137
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.91 | 16 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.5E-76 | 19 | 137 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.4E-50 | 22 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 6.6E-5 | 436 | 523 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.71E-16 | 437 | 523 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.5E-4 | 437 | 510 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 1.71E-16 | 624 | 730 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-17 | 629 | 731 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.14E-13 | 630 | 728 | No hit | No description |
| PROSITE profile | PS50297 | 17.263 | 636 | 730 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.4E-7 | 642 | 731 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.055 | 669 | 698 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.618 | 669 | 701 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 670 | 708 | 737 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 4.2 | 842 | 864 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.718 | 843 | 872 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 2.12E-7 | 844 | 893 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.0056 | 844 | 863 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0096 | 865 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 866 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.4E-4 | 868 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1032 aa Download sequence Send to blast |
MAEVRKYGLP NQPPDIPQIL LEAQNRWLRP TEICHILSNY KKFSIAPEPP NRPASGSLFL 60 FDRKILRYFR KDGHNWRKKK DGKTVKEAHE KLKVGSVDVL HCYYAHGEEN ENFQRRTYWL 120 LEEGFMNIVL VHYLEVKGGK QSFSRSKEAE ESAGLSNADS PACSNSFASQ SQVASQSMDA 180 ESPISGQISE YEDAETDNCR ASSRYHPFVE MQQPVDGVMM NNMLGVSAPR YHGEMQTTTA 240 NSDNHFATHY DIAGVFNEAG AGLRGVSKTL HDSVRFAEPY PECSAEFMEP ALYSSNATME 300 SNNLDDNSRL ETFMSEALYT NNLTQKEADA LSAAGIMSSQ AENNSYTDGI RYPLLKQSSL 360 DLFKIEPDGL KKFDSFSRWM SSELPEVADL DIKSSSDAFW SSTETVNVAD GTSIPINEQL 420 DAFAVSPSLS QDQLFSIIDV SPSYACTGSR NKVLITGTFL ANKEHVENCK WSCMFGDVEV 480 PAEVLAHGSL RCYTPVHLSG RVPFYVTCSN RVACSEVREF EFRDSDARQM DTSDPQTTGI 540 NEMHLHIRLE KLLSLGPDDY EKYVMSDGKE KSEIINTISS LMLDDKCLNQ AVPLDEKEVS 600 TARDQNIEKL VKEKLYCWLV HKVHDEDKGP NVLGKEGQGV IHLVAALGYD WAVRPIITAG 660 VKVNFRDARG WTALHWAASC GRERTVGALI ANGAESGLLT DPTPQFPAGR TAADLASENG 720 HKGIAGFLAE SALTSHLSAL TLKESKDGNV KEICGLGGAE DFAESSSAQL AYRDSQAESL 780 KDSLSAVRKS TQAAARIFQA FRVESFHRKK VVEYGDDDCG LSDERTLSLV SIKNAKPGQN 840 DGSHSAAVRI QNKFRGWKGR KEFMIIRQKI VKIQAHVRGH QVRKSYRRIV WSVGIVEKII 900 LRWRRKRRGL RGFQPVKQLE GPSPIQQLEG PSQIQPAKEE EEDEYDYLKD GRKQAEGRLQ 960 RALARVKSMT QYPEAREQYS RIANRVTELQ EPQAMMIQDD MQSDGAIADG GDFMAELEEL 1020 CGDGDAPMPT IL |
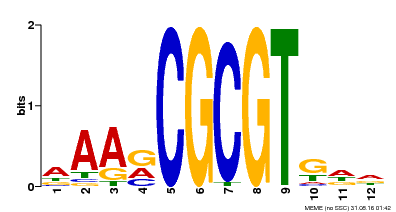
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK108461 | 0.0 | AK108461.1 Oryza sativa Japonica Group cDNA clone:002-143-D08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647012.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| TrEMBL | A0A0E0QBG1 | 0.0 | A0A0E0QBG1_ORYRU; Uncharacterized protein | ||||
| STRING | ORUFI07G23760.2 | 0.0 | (Oryza rufipogon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.1 | 0.0 | ethylene induced calmodulin binding protein | ||||



