 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_00552A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 635aa MW: 67640.5 Da PI: 6.8354 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 90.1 | 2e-28 | 148 | 201 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av++L G++kA+Pk+ilelm+v+gLt+e+v+SHLQk+Rl
Oropetium_20150105_00552A 148 KPRVVWSVELHQQFVNAVNHL-GIDKAVPKKILELMNVPGLTRENVASHLQKFRL 201
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 50.6 | 1e-17 | 6 | 73 | 43 | 109 |
H..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 43 d..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGakdflsKpfdpeelvk 109
+ +D+i+ D+ mp+mdG++ll+ e++lp+i+++a ++ +++ + +k Ga d+l Kp+ +eel++
Oropetium_20150105_00552A 6 RrgFDVIISDVHMPDMDGFRLLELVGL-EMDLPVIMMSADASTNTVMKGIKHGACDYLIKPVRMEELRN 73
3359******************87754.558************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00156 | 1.32E-18 | 1 | 78 | No hit | No description |
| PIRSF | PIRSF036392 | 1.3E-151 | 1 | 617 | IPR017053 | Response regulator B-type, plant |
| PROSITE profile | PS50110 | 32.206 | 1 | 79 | IPR001789 | Signal transduction response regulator, receiver domain |
| Gene3D | G3DSA:3.40.50.2300 | 8.5E-28 | 2 | 103 | No hit | No description |
| SuperFamily | SSF52172 | 3.7E-23 | 3 | 87 | IPR011006 | CheY-like superfamily |
| Pfam | PF00072 | 6.7E-15 | 7 | 74 | IPR001789 | Signal transduction response regulator, receiver domain |
| SuperFamily | SSF46689 | 8.24E-20 | 145 | 205 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.7E-30 | 145 | 206 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.674 | 145 | 204 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 2.7E-25 | 148 | 201 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.0E-7 | 150 | 199 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 635 aa Download sequence Send to blast |
MLRENRRGFD VIISDVHMPD MDGFRLLELV GLEMDLPVIM MSADASTNTV MKGIKHGACD 60 YLIKPVRMEE LRNIWQHVVR KKWSGNKDHE HSDSIDDTDR NRTTNNDNEY ASSANDVGDG 120 NWKSQKKKRD KEEDEGDENG DPSSTSKKPR VVWSVELHQQ FVNAVNHLGI DKAVPKKILE 180 LMNVPGLTRE NVASHLQKFR LYLKRIAQHH AGIAHPFVPS ASSAKVAPLG GFELQALAAS 240 GQIPPQALAA LQDELLGRPT SSLALPGRDQ SSLRLASIKG NKPPGEREIA FGQPMYKCAN 300 NAYGAFPPTS STVGGMPPFA AWPSNKHSMT DSGHTLGSLN NSQNSNMLLQ ELQQQPDALL 360 SGTLHSIDVK PSGLVMPSQA LSTFPASDGL SPNQNPLVMP PQSSSFLGAV PSSMKHEPLL 420 TSSQPSSSLL GGIDMVSQAS TSQPLITTHG GNLPGLMNRS SNALSSQGIS NFQGGNNPYV 480 VANQNAVGVG SKSQGVLKTE STDSLRHNYG FVGGSSSMDS GLLSQSKSPQ FGLLQSPDDV 540 GSWSSLQNID SYGGTLGPGQ SGSTSSNFQS SNVALGKLSD QGRGRNHGFV GKGTCIPSRF 600 AVDEIESPTN NLSHSIGSSG DIGNPDIFGF SGQM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 6e-24 | 144 | 207 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
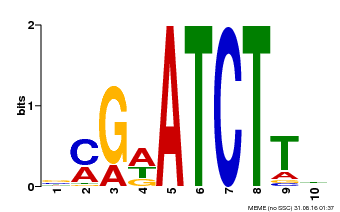
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_00552A |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002468245.1 | 0.0 | two-component response regulator ORR21 | ||||
| Refseq | XP_021306470.1 | 0.0 | two-component response regulator ORR21 | ||||
| Swissprot | A2XE31 | 0.0 | ORR21_ORYSI; Two-component response regulator ORR21 | ||||
| TrEMBL | A0A1E5WFP9 | 0.0 | A0A1E5WFP9_9POAL; Two-component response regulator ORR21 (Fragment) | ||||
| STRING | Sb01g042400.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5796 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 3e-92 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_00552A |



