 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_01978A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 327aa MW: 34075.5 Da PI: 7.3048 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61.1 | 1.8e-19 | 42 | 102 | 3 | 57 |
--SS--HHHHHHHHHHHHH..SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFek..nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
+R+t+t+eql++L+el+++ r p+ e+++++++ l +++ ++V++WFqN++a+e++
Oropetium_20150105_01978A 42 SRWTPTPEQLKILKELYYScgVRSPTSEQIQRITAMLrqhgKIEGKNVFYWFQNHKARERQ 102
7*****************7679*************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 114.3 | 5.9e-37 | 41 | 104 | 3 | 65 |
Wus_type_Homeobox 3 rtRWtPtpeQikiLeelyk.sGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
+RWtPtpeQ+kiL+ely+ +G+r+P++e+iqrita+L+++Gkie+kNVfyWFQN+kaRerqk+
Oropetium_20150105_01978A 41 GSRWTPTPEQLKILKELYYsCGVRSPTSEQIQRITAMLRQHGKIEGKNVFYWFQNHKARERQKK 104
68***************9978*****************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.13E-12 | 36 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 10.22 | 37 | 103 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.7E-7 | 39 | 107 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 8.5E-8 | 42 | 102 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 3.0E-17 | 42 | 102 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.15E-7 | 43 | 104 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0080166 | Biological Process | stomium development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MHQQQTMSAA GEGIAAGVGG RSGGGGGGGA GGGVSVCRPS GSRWTPTPEQ LKILKELYYS 60 CGVRSPTSEQ IQRITAMLRQ HGKIEGKNVF YWFQNHKARE RQKKRLTNLD VNAPAAGAGA 120 DASHLGILSL SSSSAAGATS PSTNLGMYTG NNGGASAMQL DTTAGDYRDN TAMVTDGCFL 180 QDYMGVRSTS GGHGETGAVA TPWAWFSSSH QRATTRTTLA PETLPLFPTG DDDGPRPRQG 240 GLGGEEEASR CGGTYYVPEL SSWGAAATAA TTSTTTVTIQ QQHRFYSNNY MQVPSHETHQ 300 DTAPGAGASL ELSLSSWCSP YHAGTM* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 18 | 26 | GGRSGGGGG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor required for the formation and development of tiller buds and panicles (PubMed:25697101). Required for tiller formation and female sterility (PubMed:27194802). Required for the early developmental stages of axillary meristem formation (PubMed:25841039). Plays a role in maintaining the axillary premeristem zone and in promoting the formation of the axillary meristem by promoting OSH1 expression (PubMed:25841039). Does not seem to be involved in maintenance of the shoot apical meristem (SAM) (PubMed:25841039). {ECO:0000269|PubMed:25697101, ECO:0000269|PubMed:25841039, ECO:0000269|PubMed:27194802}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00265 | DAP | Transfer from AT2G17950 | Download |
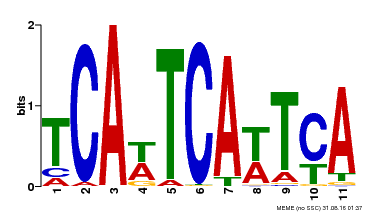
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_01978A |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by the cytokinin 6-benzylaminopurine. {ECO:0000269|PubMed:17351053}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM234744 | 8e-87 | AM234744.1 Zea mays mRNA for WUS1 protein. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022680535.1 | 1e-106 | WUSCHEL-related homeobox 1B | ||||
| Swissprot | Q7XM13 | 1e-75 | WOX1_ORYSJ; WUSCHEL-related homeobox 1 | ||||
| TrEMBL | K3ZCE4 | 1e-105 | K3ZCE4_SETIT; Uncharacterized protein | ||||
| STRING | Si024219m | 1e-106 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12465 | 31 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G17950.1 | 1e-33 | WOX family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_01978A |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



