 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_02879A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 263aa MW: 27786.8 Da PI: 8.0297 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 203.4 | 9.1e-63 | 20 | 218 | 5 | 170 |
YABBY 5 ssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakas...qllaaeshldeslkee............... 70
s +eq+Cyv+Cn C+tilav vP +slfk+vtvrCGhC++llsvnl+ + +a+++l + ++ +
Oropetium_20150105_02879A 20 SPTEQLCYVHCNCCDTILAVGVPCSSLFKTVTVRCGHCANLLSVNLRGLLlppAAPPASNQLLNFGQSSllsptsphglldela 103
578********************************************998665566777776555555578889999*888888 PP
YABBY 71 ......lleelkveeenlksnve.keesastsvssekl......senedee..vprvppvirPPekrqrvPsaynrfikeeiqr 139
l+e+ + ++++ +++s ++v++ + ++e+ e ++ +p v+rPPekrqrvPsaynrfik+eiqr
Oropetium_20150105_02879A 104 lqapslLMEQASANLSSITGGGGrSYSSCASNVPAVPMqqpvkpVHHEQPElpIKSAPSVNRPPEKRQRVPSAYNRFIKDEIQR 187
87776633333333333333222022222333333344666665333444411445677************************* PP
YABBY 140 ikasnPdishreafsaaaknWahfPkihfgl 170
ika+nPdi+hreafsaaaknWahfP+ihfgl
Oropetium_20150105_02879A 188 IKAGNPDITHREAFSAAAKNWAHFPHIHFGL 218
*****************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 7.4E-69 | 21 | 218 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 1.57E-7 | 163 | 211 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 3.6E-4 | 167 | 210 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 263 aa Download sequence Send to blast |
MMSSSSSSTA AFPLDHLAPS PTEQLCYVHC NCCDTILAVG VPCSSLFKTV TVRCGHCANL 60 LSVNLRGLLL PPAAPPASNQ LLNFGQSSLL SPTSPHGLLD ELALQAPSLL MEQASANLSS 120 ITGGGGRSYS SCASNVPAVP MQQPVKPVHH EQPELPIKSA PSVNRPPEKR QRVPSAYNRF 180 IKDEIQRIKA GNPDITHREA FSAAAKNWAH FPHIHFGLMP DQGLKKTFKS QDSAEDMLLK 240 DGLYAAAAAA AAAAVNMGVT PF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Seems to be associated with phloem cell differentiation. {ECO:0000269|PubMed:17676337}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
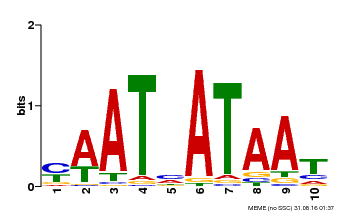
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_02879A |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002452852.1 | 1e-139 | protein YABBY 4 | ||||
| Swissprot | A2X7Q3 | 1e-124 | YAB4_ORYSI; Protein YABBY 4 | ||||
| Swissprot | Q6H668 | 1e-124 | YAB4_ORYSJ; Protein YABBY 4 | ||||
| TrEMBL | A0A1E5UN77 | 1e-139 | A0A1E5UN77_9POAL; Protein YABBY 4 | ||||
| STRING | Sb04g033590.1 | 1e-139 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1445 | 38 | 121 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 3e-64 | YABBY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_02879A |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



