 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_09313A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 421aa MW: 45394.3 Da PI: 7.8374 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102 | 3.7e-32 | 248 | 303 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
kpr++W+peLH+rF++a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Oropetium_20150105_09313A 248 KPRRCWAPELHRRFLQALQQLGGSHVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 303
79****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.364 | 245 | 305 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.22E-17 | 245 | 306 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-28 | 246 | 306 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.5E-27 | 248 | 303 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.9E-6 | 250 | 301 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 421 aa Download sequence Send to blast |
MEMDPPPPPP QSVHAAAGDR RRCREYLLAL EEERRKIQVF QRELPLCLDL VTQTIERMRS 60 QVDGVGSEET VSDHGPVLEE FMPLKPSLSL SSSEEEHDSA RDANRVVVGK KEDTAAPPPE 120 TKKATPDWLQ SVQLWSQEPL HHKVPYSLKP NFCVTELVPC KPVALKARKS GGAFQPFEKE 180 KRVEVPASST TAAAAASSAV VGEDDSGGDK AASVDDTAEK HGSDKETSKD ASNKGKDKEG 240 LSPNCNRKPR RCWAPELHRR FLQALQQLGG SHVATPKQIR ELMKVDGLTN DEVKSHLQKY 300 RLHTRRPNST VQSSSPSAAP PAPQFVVVGG IWVPXXXXXX XXXXAAAAAQ PPVQQLAADA 360 PGSASTLYAP VATLPSGSEQ QLQSVQRQPS RCSESRRSGS AGDARSASPA MSSSSQTTSA 420 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 6e-13 | 248 | 301 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-13 | 248 | 301 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-13 | 248 | 301 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-13 | 248 | 301 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-13 | 248 | 301 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that may play a role in response to nitrogen. May be involved in a time-dependent signaling for transcriptional regulation of nitrate-responsive genes. Binds specifically to the DNA sequence motif 5'-GAATC-3' or 5'-GAATATTC-3'. Represses the activity of its own promoter trough binding to these motifs. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00160 | DAP | Transfer from AT1G25550 | Download |
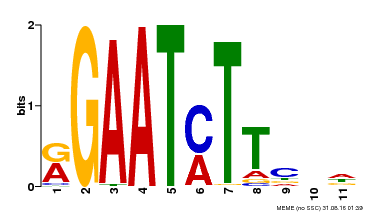
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_09313A |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by nitrate. {ECO:0000269|PubMed:23324170}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP091266 | 1e-103 | FP091266.1 Phyllostachys edulis cDNA clone: bphylf038j06, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004954868.3 | 1e-165 | transcription factor NIGT1 | ||||
| Swissprot | Q6Z869 | 1e-122 | NIGT1_ORYSJ; Transcription factor NIGT1 | ||||
| TrEMBL | A0A0A9G3F3 | 1e-172 | A0A0A9G3F3_ARUDO; Uncharacterized protein | ||||
| STRING | Si019945m | 1e-167 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6470 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 2e-62 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_09313A |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



