 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_15478A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 686aa MW: 73386 Da PI: 7.8 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.5e-16 | 454 | 500 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++Le+A+eY+k Lq
Oropetium_20150105_15478A 454 VHNLSERRRRDRINEKMRALQELIPNC-----NKIDKASMLEEAIEYLKTLQ 500
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 8.9E-21 | 447 | 509 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.38E-17 | 447 | 504 | No hit | No description |
| SuperFamily | SSF47459 | 1.15E-20 | 448 | 513 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.006 | 450 | 499 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.0E-13 | 454 | 500 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.8E-17 | 456 | 505 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 686 aa Download sequence Send to blast |
GARVWISFVS SEALSCRVAG EQEQFPATSF RTGDGIRRCI NAVVRDSNGW MGRVESSPLV 60 RRFHRNGDTS SACLATTPRV VRHPPVTTHL SPAVSPSTRM SRPTGAADDL LELLWDNGLP 120 ALRTTPSHFP PPFQPFTCSA AGSSRVHELD KQRRHDAVPP AIMEDDDXXX XXXXXXXPXX 180 XXXXXXXXXP WLHCPVIDDG DSDTAPLPPE FCAGLMMSGY SALHTAEPTE PESEPHLTAT 240 SSGIIARPQS NAAPAPSATR GAPPDTQQPK PSGGGEGVMN FTFFSRPLHW SHASAAAASP 300 GKPLESTVVQ TAAASRLRST PLFSEQRMAW LQPPKEPRAA AATPHAPPPA PTPHATTDGH 360 RRHVEAEAAA AATVTQHRLR PDARTRDATT TSSVCSGNGD GSQARPSSHQ HTGECSVSQD 420 EALIGHWLQD ADDEPSAGRR SAARGTKRSR TAEVHNLSER RRRDRINEKM RALQELIPNC 480 NKIDKASMLE EAIEYLKTLQ LQVQMMSMGS TGLCVPVPPT MLMQMQMPVV PTPMAAAAAA 540 APFPQLGGMG LGFGGMALGF AGGATQFPCP VMPGAPPMAM PPVGAKFGMP PFPHVLHGAA 600 APVEQREAAG PARPAPGGVG AGDHRRPPVP VALPVPQGMA RKDVMIGQSR AELILKPLVW 660 LVCRAGWREQ REPILPDHRE DVWLV* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 458 | 463 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
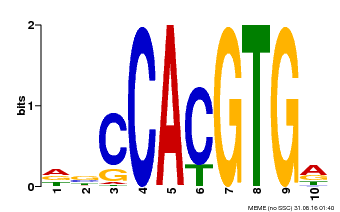
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_15478A |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB667900 | 4e-81 | AB667900.1 Oryza sativa Japonica Group APG mRNA for basic helix-loop-helix protein APG, complete cds. | |||
| GenBank | AK287958 | 4e-81 | AK287958.1 Oryza sativa Japonica Group cDNA, clone: J075053F01, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004960433.2 | 1e-120 | transcription factor APG | ||||
| TrEMBL | A0A0A9P987 | 1e-134 | A0A0A9P987_ARUDO; Uncharacterized protein | ||||
| STRING | Pavir.Ca01237.1.p | 1e-108 | (Panicum virgatum) | ||||
| STRING | GRMZM2G062541_P01 | 1e-108 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 2e-33 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_15478A |



