 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_17766A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 618aa MW: 67097.7 Da PI: 7.6069 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.3 | 1.2e-12 | 466 | 512 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
Oropetium_20150105_17766A 466 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYITDLQ 512
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 3.4E-50 | 61 | 246 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.831 | 462 | 511 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.24E-18 | 465 | 527 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.79E-15 | 465 | 516 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 5.5E-18 | 466 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.8E-10 | 466 | 512 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.9E-16 | 468 | 517 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 618 aa Download sequence Send to blast |
MVMKMEIEDD GATGGGTGGT WTEEDRALGA AVLGTDAFAY LTKGGGAISQ GLVATSLSVD 60 MQNKLQELVE SDRPGAGWNY AIYWQLSRTK SGDLVLGWGD GSCREPREGE VVAAASASND 120 DTKQRMRKRV LQRLHIAFGG ADEEDYALGI DQVTDTEIFF LASMYFAFPR RTGGPGQVFA 180 AGIPLWVPNS ERKVYPTNYC YRGFLANAAG FRTIVLVPFE NGVLELGSMQ HITESSDAVQ 240 TIRSVFSGAI DNKAAAQKPE GNGAAPTERS PSLAKIFGKD LNLGHPSAGP VVGVSKVDER 300 LWEQRSAAGG SSLPPNVQKG LQNFTWSQPR GLNSHQQKFG NGVLIVSNEA AQRSNGAADS 360 SSATQFQLQK VPPLQKLQLQ KLPQIPKTPP VVNHQQLQPQ VPRQIDFSAG SSSKPGVLVT 420 RTSVLDGESA EVDGLCKEEG PPPAVEDRRP RKRGRKPANG REEPLNHVEA ERQRREKLNQ 480 RFYALRAVVP NISKMDKASL LGDAITYITD LQKKLKEMET ERERFLESGM VGGSSTRPEV 540 DIQVVQDEVL VRVMSPMENH PVKKVFQAFE DAEVRIGESK VTGNNGTVVH SFIIKCPGAE 600 QQTREKVIAA MSRTMNS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 5e-28 | 460 | 522 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 447 | 455 | RRPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
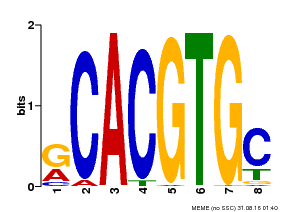
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_17766A |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025816895.1 | 0.0 | transcription factor bHLH13-like | ||||
| TrEMBL | A0A3L6SNM9 | 0.0 | A0A3L6SNM9_PANMI; Transcription factor bHLH13-like | ||||
| STRING | Pavir.Ea00030.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5174 | 38 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 2e-70 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_17766A |



