 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_19361A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 357aa MW: 38168.8 Da PI: 9.1166 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.9 | 8.1e-32 | 172 | 225 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt+ LH+rF++ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Oropetium_20150105_19361A 172 PRMRWTTSLHARFIHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 225
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-28 | 169 | 225 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.12E-15 | 170 | 226 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.7E-23 | 172 | 226 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.4E-7 | 173 | 224 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 357 aa Download sequence Send to blast |
MELFPAHPDL QLQISPSPAT KPMDLGFWKR ALDSTPVATS TAAAATSTTP PSIAIASTAP 60 TGVGFHHPTA AVHHQGAAGH LGVGALPFLH HTQPILPDSS GLRDLASMRP IRGIPVYNTS 120 QALPFLQSHP HHHHHHQHCY DAIGIGHSAG PRSPKAALRL AGAPTKRGSR APRMRWTTSL 180 HARFIHAVEL LGGHERATPK SVLELMDVKD LTLAHVKSHL QMYRTIKTTD HKPASASSYG 240 QAASKTIIDI PDDSLFDVTT TNTTSGSESS AQQSNPDGNE HGSSMCALWS NSSISRGAWF 300 HDKPSRDATP GDIKSFEDVQ SRSLDDVADL TSSPFHATGM HRSKKKPNLD FTLGPI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 4e-16 | 173 | 227 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 4e-16 | 173 | 227 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 4e-16 | 173 | 227 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 4e-16 | 173 | 227 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-16 | 173 | 227 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
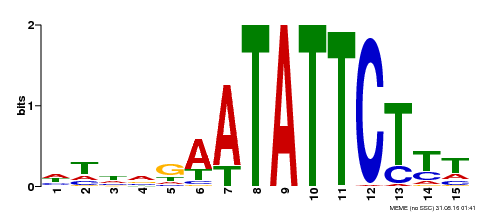
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_19361A |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT036911 | 9e-90 | BT036911.1 Zea mays full-length cDNA clone ZM_BFb0147F05 mRNA, complete cds. | |||
| GenBank | KJ726829 | 9e-90 | KJ726829.1 Zea mays clone pUT3369 G2-like transcription factor (GLK47) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025811930.1 | 0.0 | probable transcription factor KAN2 isoform X2 | ||||
| TrEMBL | A0A1E5VN24 | 0.0 | A0A1E5VN24_9POAL; Putative transcription factor KAN2 | ||||
| STRING | Pavir.Ab02797.1.p | 1e-175 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9438 | 34 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17695.1 | 1e-41 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_19361A |



