 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_21132A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 243aa MW: 26716 Da PI: 9.7508 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.1 | 9.9e-14 | 39 | 83 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT++E++++++a +++ + Wk+I + +g +t q++s+ qky
Oropetium_20150105_21132A 39 RESWTEQEHDKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 83
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.06E-17 | 33 | 89 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.408 | 34 | 88 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-7 | 37 | 89 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.0E-19 | 37 | 86 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-11 | 38 | 86 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.7E-11 | 39 | 83 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.82E-9 | 41 | 84 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
MVSAGTPPAQ AQPPQPDAAM AGDDASKKVR KPYTITKSRE SWTEQEHDKF LEALQLFDRD 60 WKKIEAFVGS KTVIQIRSHA QKYFLKVQKN GTSEHVPPPR PKRKAAHPYP QKASKNESNY 120 ALKTDSSSIH RNSGMNATVS SWPHNSIRPV VTSSMVKEDL GAGTLGPNNF CSSSTEGPPR 180 ARQPGETNDQ MNQVPSLRLM PDFTQVYSFL GSIFDPSTSG HLQKLKEMNP IDVETSGLEA 240 LV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00625 | PBM | Transfer from PK02532.1 | Download |
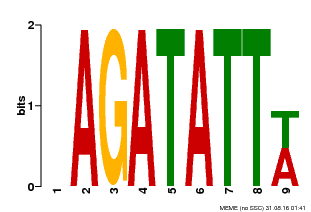
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_21132A |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP094853 | 0.0 | FP094853.1 Phyllostachys edulis cDNA clone: bphyem208j03, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002452755.1 | 1e-150 | protein REVEILLE 6 isoform X3 | ||||
| Refseq | XP_021313804.1 | 1e-150 | protein REVEILLE 6 isoform X1 | ||||
| Swissprot | Q8H0W3 | 3e-80 | RVE6_ARATH; Protein REVEILLE 6 | ||||
| TrEMBL | K3YUN1 | 1e-148 | K3YUN1_SETIT; Uncharacterized protein | ||||
| STRING | Sb04g031820.1 | 1e-149 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1096 | 38 | 111 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52660.1 | 6e-69 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_21132A |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



