 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Oropetium_20150105_24241A | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Cynodonteae; Tripogoninae; Oropetium
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 437aa MW: 46115.1 Da PI: 7.2938 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 131.6 | 2.7e-41 | 120 | 196 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
C v+gC+adls++++yhrrhkvCe+hsk+pvv+v+g+e rfCqqCsrfh l efDe+krsCr+rL++hn+rrrk+q+
Oropetium_20150105_24241A 120 CAVDGCKADLSKCRDYHRRHKVCEAHSKTPVVVVAGREMRFCQQCSRFHLLAEFDEAKRSCRKRLDGHNRRRRKPQP 196
**************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.0E-36 | 112 | 181 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.342 | 117 | 194 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 9.03E-39 | 118 | 199 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.5E-31 | 120 | 193 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 437 aa Download sequence Send to blast |
MDWDLKMPVS WDLAELEHDA VPSISVATTP VAAAASGIAA AAAAAARGAP TRAAECSVDL 60 KLGGLGEFGA GDVMKEPPAK GPAVKAXXXX XXXXXXXSVS PLKRPRSGPG GAGGSQCPSC 120 AVDGCKADLS KCRDYHRRHK VCEAHSKTPV VVVAGREMRF CQQCSRFHLL AEFDEAKRSC 180 RKRLDGHNRR RRKPQPDNMS SGSFMTGQQG TRFSSFVAPR PEASWSGIIK SEDDSYYTHQ 240 VLSSRPHFAG LTSSYSKEGR RFPFLQDGDQ VSFSAGGAAT TPLEAGTVCQ PLLKVVAAPP 300 PESSSSNKIF SPDGLTPVLD SDCALSLLSS PANSSSVDVS RMVQPTEHIP MAQPLVPSLH 360 QFGSSPGWFA ACSQAPPASS GVTAGFTCPS MESEQQLNPV LVPSSDGHEM NYHGIFHLGG 420 EGSSDGTSPP LPFSWQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-28 | 110 | 193 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 176 | 193 | KRSCRKRLDGHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
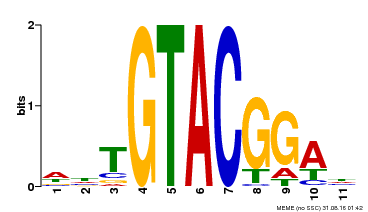
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Oropetium_20150105_24241A |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT053875 | 1e-123 | BT053875.1 Zea mays full-length cDNA clone ZM_BFb0379K14 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025803089.1 | 0.0 | squamosa promoter-binding-like protein 18 isoform X2 | ||||
| Swissprot | Q0J0K1 | 1e-162 | SPL18_ORYSJ; Squamosa promoter-binding-like protein 18 | ||||
| TrEMBL | A0A2T8KRA5 | 0.0 | A0A2T8KRA5_9POAL; Uncharacterized protein | ||||
| STRING | GRMZM2G061734_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1755 | 37 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 3e-42 | SBP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Oropetium_20150105_24241A |



