 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100054180051 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 147aa MW: 15901.1 Da PI: 10.951 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 59.2 | 5.6e-19 | 21 | 55 | 1 | 35 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkgl 35
C+ Cgt+kTplWR gp g+k+LCnaCG++ rkk++
Ote100054180051|100054180051 21 CVDCGTSKTPLWRGGPAGPKSLCNACGIRSRKKRR 55
********************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50114 | 12.202 | 15 | 51 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 1.9E-14 | 15 | 75 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 1.28E-12 | 16 | 55 | No hit | No description |
| CDD | cd00202 | 8.83E-13 | 20 | 73 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 9.4E-16 | 20 | 55 | IPR013088 | Zinc finger, NHR/GATA-type |
| Pfam | PF00320 | 8.4E-17 | 21 | 55 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 21 | 46 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 147 aa Download sequence Send to blast |
MSSQTLTPDR VSSDGSNVKT CVDCGTSKTP LWRGGPAGPK SLCNACGIRS RKKRRALMGV 60 TKDEKKAKKS CKNVISHQSQ SRNSSTVSSG DSTGKIGESS FNKKLWAFGR DVALQRPRSN 120 TNRRRKLGEE EQAAFLLMAL SCGSVYA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00335 | DAP | Transfer from AT3G06740 | Download |
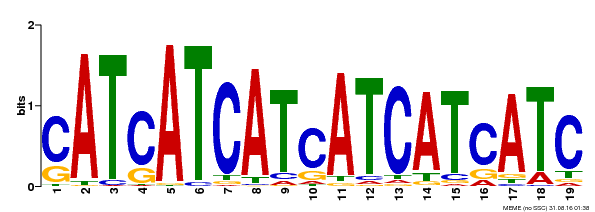
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011100810.1 | 8e-73 | GATA transcription factor 15-like | ||||
| Swissprot | Q8LG10 | 2e-35 | GAT15_ARATH; GATA transcription factor 15 | ||||
| TrEMBL | A0A4D9A7J8 | 3e-71 | A0A4D9A7J8_SALSN; Uncharacterized protein | ||||
| STRING | Migut.G00106.1.p | 4e-67 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1999 | 23 | 62 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



