 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100061220031 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 340aa MW: 37011 Da PI: 10.0214 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 156.2 | 2.3e-48 | 8 | 89 | 3 | 84 |
DUF822 3 sgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaags 84
+ r+ptwkErEnnkrRERrRRai aki++GLRaqGnyklpk++DnneVlkALcreAGw+ve+DGttyr+ + +++ ++g+
Ote100061220031|100061220031 8 AARQPTWKERENNKRRERRRRAIGAKIFTGLRAQGNYKLPKHCDNNEVLKALCREAGWIVEEDGTTYRENFVEKKHGRIVGT 89
579*****************************************************************99888777777766 PP
| |||||||
| 2 | DUF822 | 39.3 | 2.4e-12 | 106 | 175 | 71 | 144 |
DUF822 71 kgskpleeaeaagssasaspesslqsslkssalaspvesysaspksssfpspssldsislasaasllpvlsvls 144
+g+kp+ +a++ sa++s++ss+q s++ssa++sp++sy+asp+sssfpsp + d ++++ +lp+l++l+
Ote100061220031|100061220031 106 RGCKPP-QADTSMISANNSVCSSIQPSPNSSAFPSPIPSYQASPTSSSFPSPLRCDRNPTS---YILPFLRNLP 175
699**9.99999*********************************************9985...7888887765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.7E-60 | 9 | 161 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 340 aa Download sequence Send to blast |
MTACGSLAAR QPTWKERENN KRRERRRRAI GAKIFTGLRA QGNYKLPKHC DNNEVLKALC 60 REAGWIVEED GTTYRENFVE KKHGRIVGTV IQVGMLRAIT LLAELRGCKP PQADTSMISA 120 NNSVCSSIQP SPNSSAFPSP IPSYQASPTS SSFPSPLRCD RNPTSYILPF LRNLPIPTAL 180 PPLRISNSAP VTPPLSSPTR HSKPKVQLES IPASSFTSYI PLIFAASAPS SPTRRRRFAP 240 APIPERDESD ISAVNSACWV SFQTAAPTSP TYNLVKPVAQ KTSLQGDSGG EFGWGAVAKR 300 VRGSDFEFEN GAVKAWEGEK IHEIGMDDLE LTLGSGKAHA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 2e-22 | 10 | 76 | 372 | 438 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 2e-22 | 10 | 76 | 372 | 438 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 2e-22 | 10 | 76 | 372 | 438 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 2e-22 | 10 | 76 | 372 | 438 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00073 | ChIP-chip | Transfer from AT1G19350 | Download |
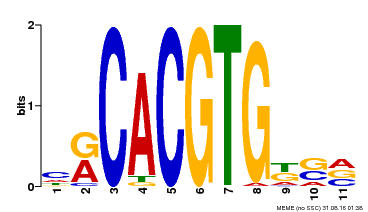
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011097209.1 | 1e-136 | BES1/BZR1 homolog protein 2-like | ||||
| Swissprot | Q94A43 | 3e-88 | BEH2_ARATH; BES1/BZR1 homolog protein 2 | ||||
| TrEMBL | A0A4D9C363 | 1e-158 | A0A4D9C363_SALSN; Brassinosteroid resistant 1/2 | ||||
| STRING | Migut.H00192.1.p | 1e-126 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA9486 | 21 | 28 |



