 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ote100093170021 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Ocimeae; Ocimum
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 232aa MW: 26757.3 Da PI: 8.0954 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 82.4 | 2.9e-26 | 11 | 52 | 10 | 51 |
HHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 10 qvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
qvtf+kRrng+lKKA+ELSvLCdaeva+iifs++gklye++s
Ote100093170021|100093170021 11 QVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCS 52
9***************************************96 PP
| |||||||
| 2 | K-box | 92.3 | 8.6e-31 | 79 | 166 | 11 | 98 |
K-box 11 eakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeen 91
ea + s+qqe+ kLk+++e Lqr+qR+llGedL++L+ keL++Le+qL+ slk+iRs++++ +l+ ++elq+ke++l+e+n
Ote100093170021|100093170021 79 EALELSSQQEYLKLKSRYEALQRTQRNLLGEDLGPLNSKELESLERQLDMSLKQIRSTRTQAMLDTLQELQRKEHALNEAN 159
4555689************************************************************************** PP
K-box 92 kaLrkkl 98
++L+++l
Ote100093170021|100093170021 160 RSLKQRL 166
****987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 23.829 | 1 | 54 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 4.6E-22 | 2 | 53 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 8.24E-26 | 10 | 69 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 8.88E-34 | 11 | 69 | No hit | No description |
| Pfam | PF00319 | 9.9E-22 | 11 | 50 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.1E-19 | 16 | 31 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.1E-19 | 31 | 52 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 14.792 | 82 | 172 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 8.1E-25 | 84 | 166 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0048833 | Biological Process | specification of floral organ number | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 232 aa Download sequence Send to blast |
MKEQLAWRHV QVTFAKRRNG LLKKAYELSV LCDAEVALII FSNRGKLYEF CSSSSMLKTL 60 ERYQKCNYGA PEPNVSTREA LELSSQQEYL KLKSRYEALQ RTQRNLLGED LGPLNSKELE 120 SLERQLDMSL KQIRSTRTQA MLDTLQELQR KEHALNEANR SLKQRLIEGS QSIHWNPNEH 180 DGVAYGRQHQ PPADHVFYHP LELEPTLQIG FHQNDPAAGP STVNNYISPW LP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 7e-45 | 68 | 169 | 1 | 104 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 7e-45 | 68 | 169 | 1 | 104 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 7e-45 | 68 | 169 | 1 | 104 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 7e-45 | 68 | 169 | 1 | 104 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00605 | ChIP-seq | Transfer from AT1G24260 | Download |
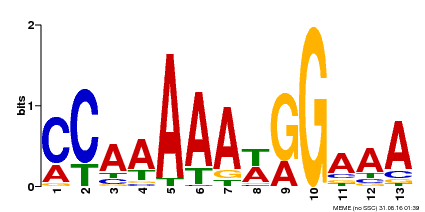
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011086825.1 | 1e-130 | agamous-like MADS-box protein AGL9 homolog | ||||
| Swissprot | Q03489 | 1e-127 | AGL9_PETHY; Agamous-like MADS-box protein AGL9 homolog | ||||
| TrEMBL | Q38733 | 1e-128 | Q38733_ANTMA; DEFH200 protein | ||||
| STRING | Solyc05g015750.2.1 | 1e-126 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



